| Sequence ID | dm3.chr3L |
|---|---|
| Location | 13,321,120 – 13,321,218 |
| Length | 98 |
| Max. P | 0.988032 |

| Location | 13,321,120 – 13,321,218 |
|---|---|
| Length | 98 |
| Sequences | 7 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.65 |
| Shannon entropy | 0.43109 |
| G+C content | 0.45001 |
| Mean single sequence MFE | -31.72 |
| Consensus MFE | -16.31 |
| Energy contribution | -16.54 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.08 |
| SVM RNA-class probability | 0.981696 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
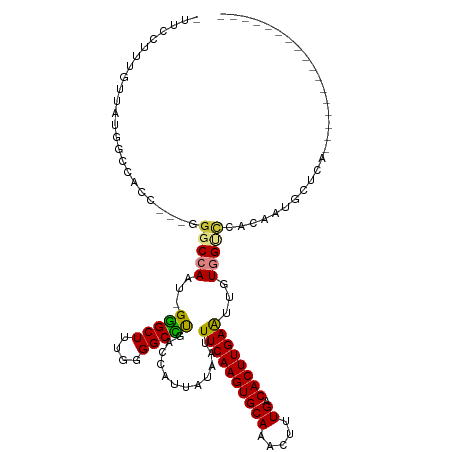
>dm3.chr3L 13321120 98 + 24543557 -----------------UGAGCAUUGUGGACCACAAUUCAAGUGUCAAAGUUUGCACUUGAAAUUAUAAUGGUCAGGCCCCAAAGCCC-AUUGGCCC---GGUGGCCAUAACAAAGGAA- -----------------......((((((.((((..(((((((((........)))))))))........((((((((......))).-..))))).---.))))))))))........- ( -32.30, z-score = -2.12, R) >droSim1.chr3L 12719097 98 + 22553184 -----------------CGAGCAUUGUGGACCACAAUUCAAGUGUCAAAGUUUGCACUUGAAAUUAUAAUGGUCAGGCCCCAAAGCCC-AUUGGCCC---GGUGGCCAUAACAAAGGAA- -----------------......((((((.((((..(((((((((........)))))))))........((((((((......))).-..))))).---.))))))))))........- ( -32.30, z-score = -2.32, R) >droSec1.super_0 5495680 98 + 21120651 -----------------UGAGCAUUGUGGACCACAAUUCAAGUGUCAAAGUUUGCACUUGAAAUUAUAAUGGUCAGGCCCCAAAGCCC-AUUGGCCU---GGUGGCCAUAACAAAGGAA- -----------------......((((((.((((..(((((((((........)))))))))...........((((((.........-...)))))---)))))))))))........- ( -33.20, z-score = -2.26, R) >droYak2.chr3L 13416788 101 + 24197627 -----------------UGAGCAUUGUGGGCCACAAUUCAAGUGUCAAAGUUUGCACUUGAAAUUAUAAUGGUUAAGCCCCAAAGCCC-AUUGGCCCAUUGCUGCCUAUAACAAAGGAA- -----------------..((((..(((((((....(((((((((........))))))))).....(((((....((......))))-)))))))))))))).(((.......)))..- ( -33.00, z-score = -2.66, R) >droEre2.scaffold_4784 13331620 99 + 25762168 -----------------UCAGCGUUGUGCGCCACCAUUCAAGUGUCAAAGUUUGCACUUGAAAUUAUAAUGGUCGGGCCCCAAAGCCCCAUUGGCCC---GGUGCCUAUAACAAAGGAA- -----------------.....(((((((((((((((((((((((........)))))))).......))))).(((((.............)))))---)))))..))))).......- ( -33.23, z-score = -2.40, R) >dp4.chrXR_group6 7723332 108 - 13314419 CAGCCAUUGUUGGCACGAAAAUUUCCCUACCAACAACUCAAGUGUCAAAAUUUGCACUUGAAACUAUAAUAGCCAAGCCCCUGAGCUC-AUUGGCC-----------ACAACAAUGGAAU ...(((((((((.........................((((((((........))))))))..........(((((((......))).-..)))).-----------.)))))))))... ( -29.00, z-score = -3.39, R) >droPer1.super_47 512244 108 - 592741 CAGCCAUUGUUGGCACGAAAAUUUCCCUACCAACAACUCAAGUGUCAAAAUUUGCACUUGAAACUAUAAUAGCCAAGCCCCUGAGCUC-AUUGGCC-----------ACAACAAUGGAAU ...(((((((((.........................((((((((........))))))))..........(((((((......))).-..)))).-----------.)))))))))... ( -29.00, z-score = -3.39, R) >consensus _________________UGAGCAUUGUGGACCACAAUUCAAGUGUCAAAGUUUGCACUUGAAAUUAUAAUGGUCAGGCCCCAAAGCCC_AUUGGCCC___GGUGGCCAUAACAAAGGAA_ ..............................((....(((((((((........)))))))))........((((((((......)))....)))))...................))... (-16.31 = -16.54 + 0.23)


| Location | 13,321,120 – 13,321,218 |
|---|---|
| Length | 98 |
| Sequences | 7 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.65 |
| Shannon entropy | 0.43109 |
| G+C content | 0.45001 |
| Mean single sequence MFE | -35.17 |
| Consensus MFE | -18.87 |
| Energy contribution | -18.53 |
| Covariance contribution | -0.34 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.30 |
| SVM RNA-class probability | 0.988032 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 13321120 98 - 24543557 -UUCCUUUGUUAUGGCCACC---GGGCCAAU-GGGCUUUGGGGCCUGACCAUUAUAAUUUCAAGUGCAAACUUUGACACUUGAAUUGUGGUCCACAAUGCUCA----------------- -.....((((...((((.((---(((((...-.))).)))))))).((((((......((((((((((.....)).))))))))..)))))).))))......----------------- ( -34.50, z-score = -2.63, R) >droSim1.chr3L 12719097 98 - 22553184 -UUCCUUUGUUAUGGCCACC---GGGCCAAU-GGGCUUUGGGGCCUGACCAUUAUAAUUUCAAGUGCAAACUUUGACACUUGAAUUGUGGUCCACAAUGCUCG----------------- -.....((((...((((.((---(((((...-.))).)))))))).((((((......((((((((((.....)).))))))))..)))))).))))......----------------- ( -34.50, z-score = -2.94, R) >droSec1.super_0 5495680 98 - 21120651 -UUCCUUUGUUAUGGCCACC---AGGCCAAU-GGGCUUUGGGGCCUGACCAUUAUAAUUUCAAGUGCAAACUUUGACACUUGAAUUGUGGUCCACAAUGCUCA----------------- -.....((((...((((.((---(((((...-.))).)))))))).((((((......((((((((((.....)).))))))))..)))))).))))......----------------- ( -35.30, z-score = -3.03, R) >droYak2.chr3L 13416788 101 - 24197627 -UUCCUUUGUUAUAGGCAGCAAUGGGCCAAU-GGGCUUUGGGGCUUAACCAUUAUAAUUUCAAGUGCAAACUUUGACACUUGAAUUGUGGCCCACAAUGCUCA----------------- -.............((((....(((((((.(-(((((....))))))...........((((((((((.....)).))))))))...)))))))...))))..----------------- ( -32.00, z-score = -1.77, R) >droEre2.scaffold_4784 13331620 99 - 25762168 -UUCCUUUGUUAUAGGCACC---GGGCCAAUGGGGCUUUGGGGCCCGACCAUUAUAAUUUCAAGUGCAAACUUUGACACUUGAAUGGUGGCGCACAACGCUGA----------------- -...(..((((...(((.((---(((((.....))).)))).)))((.(((((((....(((((((((.....)).))))))))))))))))...))))..).----------------- ( -29.90, z-score = -0.59, R) >dp4.chrXR_group6 7723332 108 + 13314419 AUUCCAUUGUUGU-----------GGCCAAU-GAGCUCAGGGGCUUGGCUAUUAUAGUUUCAAGUGCAAAUUUUGACACUUGAGUUGUUGGUAGGGAAAUUUUCGUGCCAACAAUGGCUG ...(((((((.((-----------(((((..-.((((....))))))))))).))))).(((((((((.....)).)))))))((((((((((.(((....))).))))))))))))... ( -40.00, z-score = -3.84, R) >droPer1.super_47 512244 108 + 592741 AUUCCAUUGUUGU-----------GGCCAAU-GAGCUCAGGGGCUUGGCUAUUAUAGUUUCAAGUGCAAAUUUUGACACUUGAGUUGUUGGUAGGGAAAUUUUCGUGCCAACAAUGGCUG ...(((((((.((-----------(((((..-.((((....))))))))))).))))).(((((((((.....)).)))))))((((((((((.(((....))).))))))))))))... ( -40.00, z-score = -3.84, R) >consensus _UUCCUUUGUUAUGGCCACC___GGGCCAAU_GGGCUUUGGGGCCUGACCAUUAUAAUUUCAAGUGCAAACUUUGACACUUGAAUUGUGGUCCACAAUGCUCA_________________ ........................(((((...(((((....)))))............((((((((((.....)).))))))))...)))))............................ (-18.87 = -18.53 + -0.34)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:23:18 2011