
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 13,275,342 – 13,275,435 |
| Length | 93 |
| Max. P | 0.848442 |

| Location | 13,275,342 – 13,275,435 |
|---|---|
| Length | 93 |
| Sequences | 10 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 83.00 |
| Shannon entropy | 0.34338 |
| G+C content | 0.39531 |
| Mean single sequence MFE | -23.35 |
| Consensus MFE | -11.48 |
| Energy contribution | -11.60 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.848442 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 13275342 93 - 24543557 ----GACUCCUCCACGCUUUUCACGCUUUU---CACAUUG--CCCUGGCAUGUUUUGUGUUUUAUUAGCGUGAAAAUUUUUCAAAGGUGUCAGCACUUUUUA ----(((.(((......((((((((((...---((((.((--(....))).....)))).......))))))))))........))).)))........... ( -25.44, z-score = -3.76, R) >droSim1.chr3L 12673874 93 - 22553184 ----GACUCCUCCACGCUUUUCACGCUUUU---CACAUUG--CCCUGGCAUGUUUUGUGUUUUAUUAGCGUGAAAAUUUUUCAAAGGUGUCAGCUCUUUUUA ----(((.(((......((((((((((...---((((.((--(....))).....)))).......))))))))))........))).)))........... ( -25.44, z-score = -3.98, R) >droSec1.super_0 5450547 93 - 21120651 ----GACUCCUCCACGCUUUUCACGCUUUU---CACAUUG--CCCUGGCAUGUUUUGUGUUUUAUUAGCGUGAAAAUUUUUCAAAGGUGUCAGCACUUUAUA ----(((.(((......((((((((((...---((((.((--(....))).....)))).......))))))))))........))).)))........... ( -25.44, z-score = -3.72, R) >droYak2.chr3L 13365970 93 - 24197627 ----GACUCCUCGACGCUUUUCACGCUUUU---CACAUUG--CCCUGGCAUGUUUUGUGUUUUAUUAGCGUGAAAAUUUUUCAAAGGUGUCAGCACUUUUUA ----(((.(((.((...((((((((((...---((((.((--(....))).....)))).......))))))))))....))..))).)))........... ( -25.90, z-score = -3.52, R) >droAna3.scaffold_13337 11081510 93 + 23293914 ----GACUCCUCCACGUUUUUCACGCUUUU---CACAUUG--CUCUGGCAUGUUUUGUGUUUUAUUAGCGUGAAAAUUUUUCAAAGGUGUCAGCACUUUUUA ----(((.(((......((((((((((...---((((..(--(........))..)))).......))))))))))........))).)))........... ( -25.44, z-score = -4.05, R) >dp4.chrXR_group6 5534795 99 - 13314419 CUGGGACUCCUCCACGCUUUUCACGCUUUU---CACAUUGCUCUCUGGCAUGUUUUGUGUUUUAUUAGCGUGAAAAUUUUUCAAAGGUGUCAGCACUUUUUA ...(((....)))....((((((((((...---((((.((((....)))).....)))).......))))))))))......(((((((....))))))).. ( -25.40, z-score = -2.81, R) >droWil1.scaffold_180698 6830198 93 - 11422946 ----GGAGGAGGAGGGACUUUCACGCUUUU---CACAUUG--CUGUGGCAUGUUUUGUGUUUUAUUAGCGUGAAAAUUUUUCAAAGGUGUCAGCAGUUUUUA ----.......((((((.(((((((((...---((((..(--(........))..)))).......))))))))).))))))((((.((....)).)))).. ( -23.80, z-score = -1.51, R) >droMoj3.scaffold_6680 10026177 91 + 24764193 -----------CCACGACUCUCAUGCUAUCUCGCUCAGCUGUGACUGGCAUGUUUUGUGUUUUAUUAGCGUGAAAAUUUUUCAAAGGUGUCAGCAGUUUAUA -----------....(((.(((((((((..((((......)))).)))))))(((..((((.....))))..)))..........)).)))........... ( -20.80, z-score = -0.63, R) >droVir3.scaffold_13049 12688628 88 - 25233164 -----------UCACGCUUUUCGCGCUAUCUCCCUCAGUUCUGACUGGCAUGUUUUGUAUUUUAUUAGCGUGAAAAUGUUUCAAAGGUGUCAGCAGUUU--- -----------..(((.(((((((((((.......((((....))))(((.....))).......))))))))))))))....(((.((....)).)))--- ( -18.80, z-score = -0.85, R) >droGri2.scaffold_15110 23319196 87 - 24565398 -----------CUACGCUUUUCACGCUUU----CUCAGCUCUGACUGGCAUGUUUUGUAUUUUAUAAGCGUGAAAAUUUUUCAAAGGUGUCAGCAGUUUUUA -----------......(((((((((((.----..(((......)))(((.....))).......)))))))))))......((((.((....)).)))).. ( -17.00, z-score = -0.55, R) >consensus ____GACUCCUCCACGCUUUUCACGCUUUU___CACAUUG__CCCUGGCAUGUUUUGUGUUUUAUUAGCGUGAAAAUUUUUCAAAGGUGUCAGCACUUUUUA .................((((((((((........((........))(((.....)))........)))))))))).......................... (-11.48 = -11.60 + 0.12)

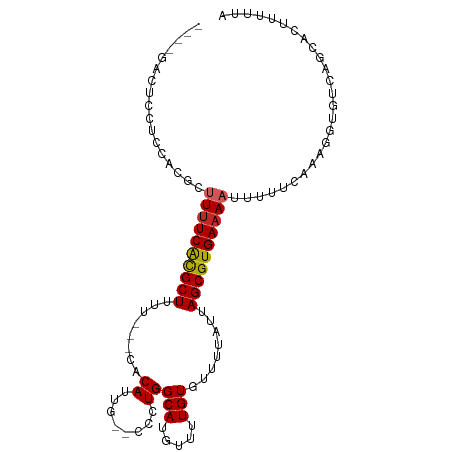

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:23:17 2011