| Sequence ID | dm3.chr3L |
|---|---|
| Location | 13,271,279 – 13,271,382 |
| Length | 103 |
| Max. P | 0.987070 |

| Location | 13,271,279 – 13,271,382 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 59.81 |
| Shannon entropy | 0.73016 |
| G+C content | 0.31308 |
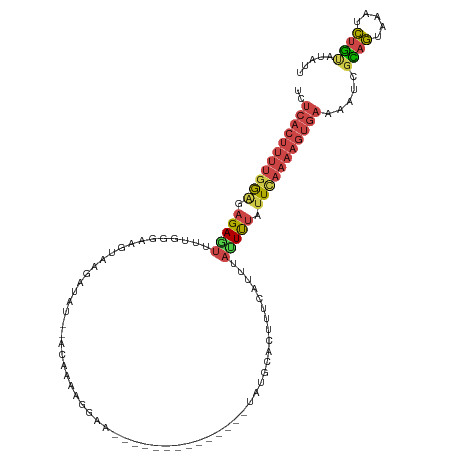
| Mean single sequence MFE | -23.21 |
| Consensus MFE | -7.73 |
| Energy contribution | -8.73 |
| Covariance contribution | 1.01 |
| Combinations/Pair | 1.50 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.33 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.26 |
| SVM RNA-class probability | 0.987070 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 13271279 103 + 24543557 UCUCACUUUUGGAGAGAAUUUUGGGAAAUACAGUAC--ACAAAGGGCC------------UAUAUUCACUUUCAUUUAUUUUCUUUAAAAGUGAAAAUCGCAGUAAAUCUGUAUAUU ..(((((((((((((((((..((..(((((.((..(--......)..)------------).))))...)..))...))))))))))))))))).....((((.....))))..... ( -25.80, z-score = -3.65, R) >droSim1.chr3L 12669843 102 + 22553184 UCUCACUUUUGGAGAGAGUUUUGGGAAGUAAGAUAU--ACAAAGGAC-------------UAUAUGCACUUUCAUUUAUUUUCUUCAAAAGUGAAAAUCGCAGUAAAUCUGUAUAUU ..(((((((((((((((((....((((((...((((--(........-------------)))))..))))))....))))))))))))))))).....((((.....))))..... ( -30.80, z-score = -4.70, R) >droSec1.super_0 5446555 92 + 21120651 UCUCACUUUUGGAGAGAGUUUUGGGAAGUAUGAUAU--A-----------------------UAUGCACUUUGAUUUAUUUUCUUCAAAAGUGAAAGUCGCAGUAAAUCUGUAUAAU ..(((((((((((((((((.(..((..(((((....--.-----------------------)))))..))..)...))))))))))))))))).....((((.....))))..... ( -26.90, z-score = -3.88, R) >droEre2.scaffold_4784 13280826 103 + 25762168 UCUCACUUUUGGAGAGAGUUUUGGGUAGUAAGCUAA--ACAUAAGGAA------------CCUAUUCGCUUUCAUUUAUUUUACUCAAAAGUGAAAACCGUAGCCAACCUGUGUAUU ...(((..((((.((((((..(((((..........--.........)------------))))...)))))).....(((((((....))))))).......))))...))).... ( -19.01, z-score = 0.10, R) >droYak2.chr3L 13361916 117 + 24197627 UCUCACUUUUGGAGAGAGUUUUCAGCAGUAAGUUAAUAAAAUAAAAAAAAGGGAUGCAAACUUAUUCGCUUUCAUUUACUUUAUUCAAAAGUGAAAAUCGUAGCCAACCUGCAUAUU ..(((((((((((.(((((....(((((((((((........................)))))))).))).......))))).))))))))))).....((((.....))))..... ( -25.46, z-score = -2.46, R) >droWil1.scaffold_180698 6821798 86 + 11422946 -----------AGGCGAACGGCGAACGGGUGCUUUUGUGUUACAAGAA--------------AGAGUACUUUAUUUUUAUCCAUUAAUUUAUAAUAUCUGUUUUAUAUAUA------ -----------..((.....))((((((((((((((...........)--------------))))))).((((..(((.....)))...))))...))))))........------ ( -11.30, z-score = 0.82, R) >consensus UCUCACUUUUGGAGAGAGUUUUGGGAAGUAAGAUAU__ACAAAAGGAA______________UAUGCACUUUCAUUUAUUUUAUUCAAAAGUGAAAAUCGCAGUAAAUCUGUAUAUU ..(((((((((((.(((((..........................................................))))).))))))))))).....((((.....))))..... ( -7.73 = -8.73 + 1.01)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:23:15 2011