| Sequence ID | dm3.chr3L |
|---|---|
| Location | 13,265,019 – 13,265,113 |
| Length | 94 |
| Max. P | 0.847441 |

| Location | 13,265,019 – 13,265,113 |
|---|---|
| Length | 94 |
| Sequences | 12 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 77.87 |
| Shannon entropy | 0.45117 |
| G+C content | 0.46058 |
| Mean single sequence MFE | -24.72 |
| Consensus MFE | -16.40 |
| Energy contribution | -16.25 |
| Covariance contribution | -0.15 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.847441 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
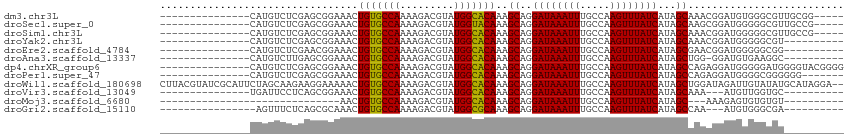
>dm3.chr3L 13265019 94 + 24543557 ---------------CAUGUCUCGAGCGGAAACUGUGCCAAAAGACGUAUGGCACAAAGCAGGAUAAAUUUGCCAAGUUUAUCAUAGCAAACGGAUGUGGGCGUUGCGG----- ---------------((((((.((.(((....)(((((((.........)))))))..))..((((((((.....))))))))........))))))))..........----- ( -25.60, z-score = -0.80, R) >droSec1.super_0 5440372 94 + 21120651 ---------------CAUGUCUCGAGCGGAAACUGUGCCAAAAGACGUAUGGUACAAAGCAGGAUAAAUUUGCCAAGUUUAUCAUAGCAAGCGGAUGGGGGCGUUGCCG----- ---------------.(((((((..(((....)(((((((.........)))))))..((..((((((((.....))))))))...))..)).....))))))).....----- ( -24.20, z-score = -0.18, R) >droSim1.chr3L 12663681 94 + 22553184 ---------------CAUGUCUCGAGCGGAAACUGUGCCAAAAGACGUAUGGCACAAAGCAGGAUAAAUUUGCCAAGUUUAUCAUAGCAAACGGAUGGGGGCGUUGCCG----- ---------------.(((((((.(.((.....(((((((.........)))))))..((..((((((((.....))))))))...))...))..).))))))).....----- ( -26.20, z-score = -0.93, R) >droYak2.chr3L 13355672 89 + 24197627 ---------------CAUGUCUCGAGCGGAAACUGUGCCAAAAGACGUAUGGCACAAAGCAGGAUAAAUUUGCCAAGUUUAUCAUAGCAAACGGAUGGGGGCGU---------- ---------------.(((((((.(.((.....(((((((.........)))))))..((..((((((((.....))))))))...))...))..).)))))))---------- ( -26.10, z-score = -2.04, R) >droEre2.scaffold_4784 13274751 89 + 25762168 ---------------CAUGUCUCGAACGGAAACUGUGCCAAAAGACGUAUGGCACAAAGCAGGAUAAAUUUGCCAAGUUUAUCAUAGCGAACGGAUGGGGGCGG---------- ---------------..((((((.(.((.....(((((((.........)))))))..((..((((((((.....))))))))...))...))..).)))))).---------- ( -25.30, z-score = -2.06, R) >droAna3.scaffold_13337 11071407 88 - 23293914 ---------------CAUGUCUUGAGCGGAAACUGUGCCAAAAGACGUAUGGCACAAAGCAGGAUAAAUUUGCCAAGUUUAUCAUAGCUGG-GGAUGUGAAGGC---------- ---------------((((((((.((((....)(((((((.........)))))))......((((((((.....))))))))...))).)-))))))).....---------- ( -29.30, z-score = -3.34, R) >dp4.chrXR_group6 5521996 99 + 13314419 ---------------CAUGUCUCGAGCGGAAACUGUGCCAAAAGACGUAUGGCACAAAGCAGGAUAAAUUUGCCAAGUUUAUCAUAGCCAGAGGAUGGGGGAUGGGGUACGGGG ---------------.....((((.(((....)(((((((.........)))))))..))..........((((..(((..((((..((...))))))..)))..)))))))). ( -26.00, z-score = -0.96, R) >droPer1.super_47 11021 92 + 592741 ---------------CAUGUCUCGAGCGGAAACUGUGCCAAAAGACGUAUGGCACAAAGCAGGAUAAAUUUGCCAAGUUUAUCAUAGCCAGAGGAUGGGGCGGGGGG------- ---------------..(((((((.(((....)(((((((.........)))))))..))..((((((((.....))))))))............))))))).....------- ( -25.40, z-score = -1.12, R) >droWil1.scaffold_180698 6814286 112 + 11422946 CUUACGUAUCGCAUUCUAGCAAGAAGGAAAAACUGUGCCAAAAGACGUAUGGCACAAAGCAGGAUAAAUUUGCCAAGUUUAUCAUAGCUGGAUAGAUUGUAUAUGCAUAGGA-- ((((.((((.(((.((((((.............(((((((.........)))))))......((((((((.....))))))))...)))))).....)))..)))).)))).-- ( -28.30, z-score = -1.93, R) >droVir3.scaffold_13049 12674616 86 + 25233164 ---------------UGAUUCCUCAGCGGAAACUGUGCCAAAAGACGUAUGGCACAAAGCAGGAUAAAUUUGCCAAGUUUAUCAUAGCAAA---AUGUUGGUGC---------- ---------------.......((((((....)(((((((.........)))))))..((..((((((((.....))))))))...))...---..)))))...---------- ( -21.80, z-score = -1.26, R) >droMoj3.scaffold_6680 10007785 71 - 24764193 ------------------------------AACUGUGCCAAAAGACGUAUGGCACAAAGCAGGAUAAAUUUGCCAAGUUUAUCAUAGC---AAAGAGUGUGUGU---------- ------------------------------...(((((((.........)))))))..((..((((((((.....))))))))...))---.............---------- ( -16.90, z-score = -1.41, R) >droGri2.scaffold_15110 23305793 85 + 24565398 ----------------AGUUUCUCAGCGCAAACUGUGCCAAAAGACGUAUGGCGCAAAGCAGGAUAAAUUUGCCAAGUUUAUCAUAGCCAA---AUGUGGGCGA---------- ----------------..........(((..(((((((((.........)))))))..((..((((((((.....))))))))...))...---..))..))).---------- ( -21.50, z-score = -0.32, R) >consensus _______________CAUGUCUCGAGCGGAAACUGUGCCAAAAGACGUAUGGCACAAAGCAGGAUAAAUUUGCCAAGUUUAUCAUAGCAAACGGAUGGGGGCGU__________ .................................(((((((.........)))))))..((..((((((((.....))))))))...)).......................... (-16.40 = -16.25 + -0.15)

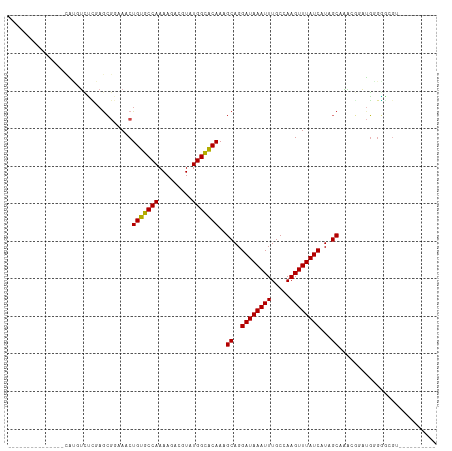
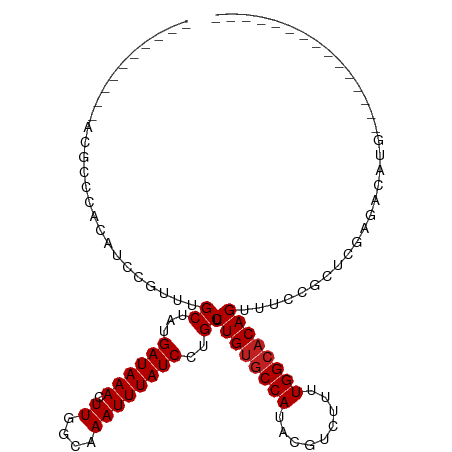
| Location | 13,265,019 – 13,265,113 |
|---|---|
| Length | 94 |
| Sequences | 12 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 77.87 |
| Shannon entropy | 0.45117 |
| G+C content | 0.46058 |
| Mean single sequence MFE | -19.35 |
| Consensus MFE | -13.48 |
| Energy contribution | -13.64 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.18 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.816854 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 13265019 94 - 24543557 -----CCGCAACGCCCACAUCCGUUUGCUAUGAUAAACUUGGCAAAUUUAUCCUGCUUUGUGCCAUACGUCUUUUGGCACAGUUUCCGCUCGAGACAUG--------------- -----.......((........((((((((.........)))))))).......))..(((((((.........)))))))(((((.....)))))...--------------- ( -21.36, z-score = -1.31, R) >droSec1.super_0 5440372 94 - 21120651 -----CGGCAACGCCCCCAUCCGCUUGCUAUGAUAAACUUGGCAAAUUUAUCCUGCUUUGUACCAUACGUCUUUUGGCACAGUUUCCGCUCGAGACAUG--------------- -----.(((((.((........))))))).......((..((((.........))))..)).......(((((..(((.........))).)))))...--------------- ( -17.00, z-score = 0.21, R) >droSim1.chr3L 12663681 94 - 22553184 -----CGGCAACGCCCCCAUCCGUUUGCUAUGAUAAACUUGGCAAAUUUAUCCUGCUUUGUGCCAUACGUCUUUUGGCACAGUUUCCGCUCGAGACAUG--------------- -----(((.(((((........((((((((.........)))))))).......))..(((((((.........)))))))))).)))...........--------------- ( -23.06, z-score = -1.48, R) >droYak2.chr3L 13355672 89 - 24197627 ----------ACGCCCCCAUCCGUUUGCUAUGAUAAACUUGGCAAAUUUAUCCUGCUUUGUGCCAUACGUCUUUUGGCACAGUUUCCGCUCGAGACAUG--------------- ----------..((........((((((((.........)))))))).......))..(((((((.........)))))))(((((.....)))))...--------------- ( -21.36, z-score = -2.20, R) >droEre2.scaffold_4784 13274751 89 - 25762168 ----------CCGCCCCCAUCCGUUCGCUAUGAUAAACUUGGCAAAUUUAUCCUGCUUUGUGCCAUACGUCUUUUGGCACAGUUUCCGUUCGAGACAUG--------------- ----------................((...((((((.((....))))))))..))..(((((((.........)))))))(((((.....)))))...--------------- ( -18.00, z-score = -1.08, R) >droAna3.scaffold_13337 11071407 88 + 23293914 ----------GCCUUCACAUCC-CCAGCUAUGAUAAACUUGGCAAAUUUAUCCUGCUUUGUGCCAUACGUCUUUUGGCACAGUUUCCGCUCAAGACAUG--------------- ----------((..........-..(((...((((((.((....))))))))..)))((((((((.........)))))))).....))..........--------------- ( -16.30, z-score = -0.78, R) >dp4.chrXR_group6 5521996 99 - 13314419 CCCCGUACCCCAUCCCCCAUCCUCUGGCUAUGAUAAACUUGGCAAAUUUAUCCUGCUUUGUGCCAUACGUCUUUUGGCACAGUUUCCGCUCGAGACAUG--------------- .....................(((.(((............((((.........))))((((((((.........)))))))).....))).))).....--------------- ( -19.50, z-score = -1.32, R) >droPer1.super_47 11021 92 - 592741 -------CCCCCCGCCCCAUCCUCUGGCUAUGAUAAACUUGGCAAAUUUAUCCUGCUUUGUGCCAUACGUCUUUUGGCACAGUUUCCGCUCGAGACAUG--------------- -------..............(((.(((............((((.........))))((((((((.........)))))))).....))).))).....--------------- ( -19.50, z-score = -1.32, R) >droWil1.scaffold_180698 6814286 112 - 11422946 --UCCUAUGCAUAUACAAUCUAUCCAGCUAUGAUAAACUUGGCAAAUUUAUCCUGCUUUGUGCCAUACGUCUUUUGGCACAGUUUUUCCUUCUUGCUAGAAUGCGAUACGUAAG --.....(((((........((((.......))))...(((((((............((((((((.........))))))))..........))))))).)))))......... ( -20.55, z-score = -0.72, R) >droVir3.scaffold_13049 12674616 86 - 25233164 ----------GCACCAACAU---UUUGCUAUGAUAAACUUGGCAAAUUUAUCCUGCUUUGUGCCAUACGUCUUUUGGCACAGUUUCCGCUGAGGAAUCA--------------- ----------(((.......---(((((((.........))))))).......)))..(((((((.........)))))))(.((((.....)))).).--------------- ( -22.74, z-score = -2.28, R) >droMoj3.scaffold_6680 10007785 71 + 24764193 ----------ACACACACUCUUU---GCUAUGAUAAACUUGGCAAAUUUAUCCUGCUUUGUGCCAUACGUCUUUUGGCACAGUU------------------------------ ----------..........(((---((((.........)))))))...........((((((((.........))))))))..------------------------------ ( -16.10, z-score = -2.29, R) >droGri2.scaffold_15110 23305793 85 - 24565398 ----------UCGCCCACAU---UUGGCUAUGAUAAACUUGGCAAAUUUAUCCUGCUUUGCGCCAUACGUCUUUUGGCACAGUUUGCGCUGAGAAACU---------------- ----------..(((.....---..))).........((..((...........((.(((.((((.........)))).)))...))))..)).....---------------- ( -16.70, z-score = 0.46, R) >consensus __________ACGCCCACAUCCGUUUGCUAUGAUAAACUUGGCAAAUUUAUCCUGCUUUGUGCCAUACGUCUUUUGGCACAGUUUCCGCUCGAGACAUG_______________ ..........................((...((((((.((....))))))))..)).((((((((.........))))))))................................ (-13.48 = -13.64 + 0.17)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:23:13 2011