| Sequence ID | dm3.chr3L |
|---|---|
| Location | 13,257,318 – 13,257,413 |
| Length | 95 |
| Max. P | 0.779907 |

| Location | 13,257,318 – 13,257,413 |
|---|---|
| Length | 95 |
| Sequences | 9 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 81.05 |
| Shannon entropy | 0.36178 |
| G+C content | 0.45188 |
| Mean single sequence MFE | -29.30 |
| Consensus MFE | -21.03 |
| Energy contribution | -22.30 |
| Covariance contribution | 1.27 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.779907 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 13257318 95 - 24543557 -----------------------CAUACAUGGCAAGUGUUGUAAGUAUGUGCUUAAAUACGCAUUUAUUCAUACAAAUAUGGCCAGAAGGCGUCUGCGGCAACCGCAGAAAGCAUCAG -----------------------......((((.....(((((((((.((((........)))).))))..))))).....))))((..((.(((((((...)))))))..)).)).. ( -28.20, z-score = -2.27, R) >droSim1.chr3L 12655047 109 - 22553184 ---------UCGGGCUCGGGCUCCAUACAUGGCAAGUGUUGUAAGUAUGUGCUUAAAUACGCAUUUAUUCAUACAAAUAUGGCCAGAAGGCGUCUGCGGCAACCGCAGAAAGCAUCAG ---------..((((....)).)).....((((.....(((((((((.((((........)))).))))..))))).....))))((..((.(((((((...)))))))..)).)).. ( -32.60, z-score = -1.36, R) >droSec1.super_0 5432230 97 - 21120651 ---------------------UCCAUACAUGGCAAGUGUUGUAAGUAUGUGCUUAAAUACGCAUUUAUUCAUACAAAUAUGGCCAGAAGGCGUCUGCGGCAACCGCAGAAAGCAUCAG ---------------------........((((.....(((((((((.((((........)))).))))..))))).....))))((..((.(((((((...)))))))..)).)).. ( -28.20, z-score = -2.15, R) >droYak2.chr3L 13347289 117 - 24197627 -GCUGGCUCUCGGGCUGGGGCUCCAUACAUGGCAAGUGUUGUAAGUAUGUGCUUAAAUACGCAUUUAUUCAUACAAAUAUGGCCAGAAGGCGUCUGCGGCAACCGCAGAAAGCAUCAG -(((.((..((((((....)).)).....((((.....(((((((((.((((........)))).))))..))))).....))))))..)).(((((((...))))))).)))..... ( -39.00, z-score = -1.45, R) >droEre2.scaffold_4784 13266630 118 - 25762168 GGCCCGGACUCGGGCUCGGUCUCCAUACAUGGCAAGUGUUGUAAGUAUGUGCUUAAAUACGCAUUUAUUCAUACAAAUAUGGCCAGAAGGCGUCUGCGGCAACCGCAGAAAGCAUCAG ((((((....)))))).............((((.....(((((((((.((((........)))).))))..))))).....))))((..((.(((((((...)))))))..)).)).. ( -37.90, z-score = -1.66, R) >droPer1.super_47 2298 98 - 592741 --------------------CACCAUACAUGGCAAGUGUUGUAAGUAUGUGCUUAAAUACGCAUUUAUUCAUACAAAUAUGGCCAGAAGGCGUCUGCGGCAACCGCAGAAAGCAUCAG --------------------.........((((.....(((((((((.((((........)))).))))..))))).....))))((..((.(((((((...)))))))..)).)).. ( -28.20, z-score = -2.00, R) >dp4.chrXR_group6 5513136 98 - 13314419 --------------------CACCAUACAUGGCAAGUGUUGUAAGUAUGUGCUUAAAUACGCAUUUAUUCAUACAAAUAUGGCCAGAAGGCGUCUGCGGCAACCGCAGAAAGCAUCAG --------------------.........((((.....(((((((((.((((........)))).))))..))))).....))))((..((.(((((((...)))))))..)).)).. ( -28.20, z-score = -2.00, R) >droWil1.scaffold_180698 6803638 89 - 11422946 -----------------------CAUAGAUGGCAAGUGUUGUAAGUAUGUGCUUAAAUACGCAUUUAUUCAUACAAAUAUGGCCAGGAGGCGGCGGCUGGCUAUGGCAACAG------ -----------------------.............((((((..(((((....(((((....)))))..)))))...(((((((((..(....)..))))))))))))))).------ ( -24.00, z-score = -0.68, R) >droVir3.scaffold_13049 12665139 77 - 25233164 -----------------------UGUACAUGGCAAGUGUUGUAAGUAUGUGCUUAAAUACGCAUUUAUUCAUACAAAUAUGGCCA-ACGGCAGCAGCAGGA----------------- -----------------------(((...((((.....(((((((((.((((........)))).))))..))))).....))))-...))).........----------------- ( -17.40, z-score = 0.22, R) >consensus ____________________CUCCAUACAUGGCAAGUGUUGUAAGUAUGUGCUUAAAUACGCAUUUAUUCAUACAAAUAUGGCCAGAAGGCGUCUGCGGCAACCGCAGAAAGCAUCAG .............................((((.....(((((((((.((((........)))).))))..))))).....))))....((.((((((.....))))))..))..... (-21.03 = -22.30 + 1.27)


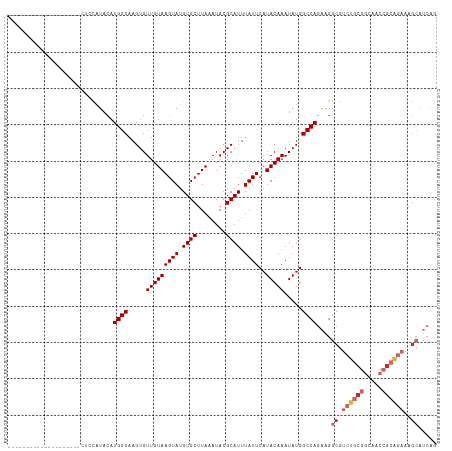
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:23:10 2011