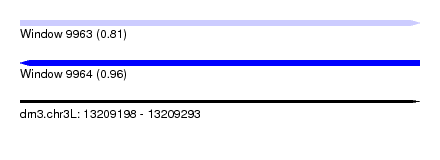
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 13,209,198 – 13,209,293 |
| Length | 95 |
| Max. P | 0.963527 |

| Location | 13,209,198 – 13,209,293 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 92.00 |
| Shannon entropy | 0.15216 |
| G+C content | 0.53460 |
| Mean single sequence MFE | -28.48 |
| Consensus MFE | -24.16 |
| Energy contribution | -23.38 |
| Covariance contribution | -0.77 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.85 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.807672 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
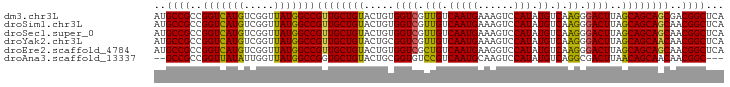
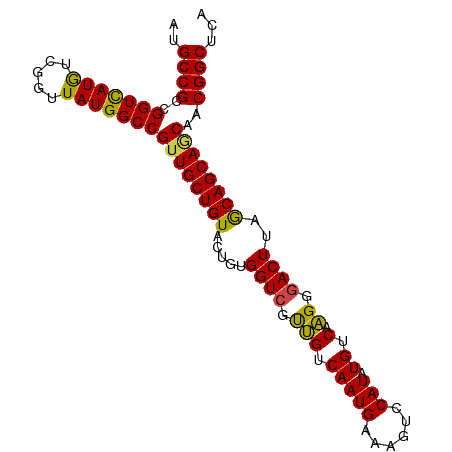
>dm3.chr3L 13209198 95 + 24543557 UGAGCCGUCGCUGCUGCUAAGUCCCUUGACAUAUGGACUUUCAUUGACAACGACCACAGUACAGCAACGGCCAUAACCGACAUGACCGGCGGCAU ...((((((((((.((((((((((..........))))))...(((....)))....))))))))..(((......)))........)))))).. ( -28.40, z-score = -2.08, R) >droSim1.chr3L 12604531 95 + 22553184 UGAGCCGUUGCUGCUGCUAAGUCCCUUGACAUAUGGACUUUCAUUGACAACGACCACAGUACAGCAACGGCCAUAACCGACAUGACCGGCGGCAU ((.((((((((((.((((((((((..........))))))...(((....)))....))))))))))))))))...(((.(......).)))... ( -29.30, z-score = -2.51, R) >droSec1.super_0 5384662 95 + 21120651 UGAGCCGUUGCUGCUGCUAAGUCCCUUGACAUAUGGACUUUCAUUGACAACGACCACAGUACAGCAACGGCCAUAACCGACAUGACCGGCGGCAU ((.((((((((((.((((((((((..........))))))...(((....)))....))))))))))))))))...(((.(......).)))... ( -29.30, z-score = -2.51, R) >droYak2.chr3L 13296558 95 + 24197627 UGAGCCGUUGUUGCUGCUAAGUCCCUUGACAUAUGGACUUUCAUUGACAACGACCGCAGUACAGCAACGGCCAUAACCGACAUGACCGGCGGCAU ((.((((((((((((((.((((((..........))))))...(((....)))..))))..))))))))))))...(((.(......).)))... ( -28.70, z-score = -1.80, R) >droEre2.scaffold_4784 13219085 95 + 25762168 UGAGCCGUUGCUGCUGCUAAGUCCCUUGACAUAUGGACCUUCAUUGACAGCGACCACAGUACAGCAACGGCCAUAACCGACAUGACCGGCGGCAU ((.((((((((((.((((..(((.((.(.((.((((....)))))).))).)))...))))))))))))))))...(((.(......).)))... ( -29.20, z-score = -1.83, R) >droAna3.scaffold_13337 11021423 90 - 23293914 ---GCCGUUGUUGCUGUUAAGUCGCCUGACAUAUGGACUUGCAUUGACGGACACCGCAGUACAGCACCGGCCAUAACCAAUAUAACCGGCGGC-- ---(((((((.((((((((((((............)))))).((((.(((...)))))))))))))..((......))........)))))))-- ( -26.00, z-score = -0.31, R) >consensus UGAGCCGUUGCUGCUGCUAAGUCCCUUGACAUAUGGACUUUCAUUGACAACGACCACAGUACAGCAACGGCCAUAACCGACAUGACCGGCGGCAU ...((((((((((.((((..(..(.(((.((.(((......))))).))).)..)..)))))))))))))).....(((.(......).)))... (-24.16 = -23.38 + -0.77)

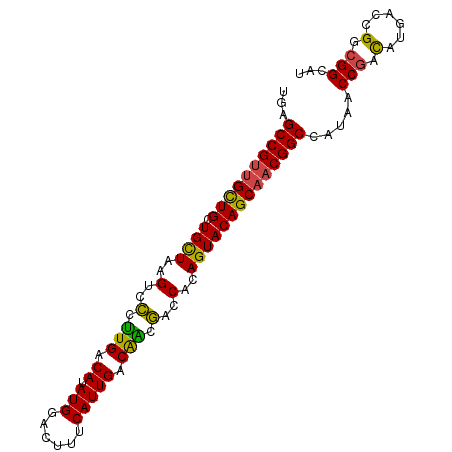
| Location | 13,209,198 – 13,209,293 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 92.00 |
| Shannon entropy | 0.15216 |
| G+C content | 0.53460 |
| Mean single sequence MFE | -35.30 |
| Consensus MFE | -33.67 |
| Energy contribution | -32.98 |
| Covariance contribution | -0.69 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.95 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.72 |
| SVM RNA-class probability | 0.963527 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 13209198 95 - 24543557 AUGCCGCCGGUCAUGUCGGUUAUGGCCGUUGCUGUACUGUGGUCGUUGUCAAUGAAAGUCCAUAUGUCAAGGGACUUAGCAGCAGCGACGGCUCA ..((((.((((((((.....)))))))((((((((.......((((.....))))((((((..........)))))).))))))))).))))... ( -36.20, z-score = -2.04, R) >droSim1.chr3L 12604531 95 - 22553184 AUGCCGCCGGUCAUGUCGGUUAUGGCCGUUGCUGUACUGUGGUCGUUGUCAAUGAAAGUCCAUAUGUCAAGGGACUUAGCAGCAGCAACGGCUCA .....((((((((((.....)))))))(((((((..((((..((((.....))))((((((..........)))))).))))))))))))))... ( -35.90, z-score = -2.24, R) >droSec1.super_0 5384662 95 - 21120651 AUGCCGCCGGUCAUGUCGGUUAUGGCCGUUGCUGUACUGUGGUCGUUGUCAAUGAAAGUCCAUAUGUCAAGGGACUUAGCAGCAGCAACGGCUCA .....((((((((((.....)))))))(((((((..((((..((((.....))))((((((..........)))))).))))))))))))))... ( -35.90, z-score = -2.24, R) >droYak2.chr3L 13296558 95 - 24197627 AUGCCGCCGGUCAUGUCGGUUAUGGCCGUUGCUGUACUGCGGUCGUUGUCAAUGAAAGUCCAUAUGUCAAGGGACUUAGCAGCAACAACGGCUCA ..((((..(((((((.....)))))))((((((((.......((((.....))))((((((..........)))))).))))))))..))))... ( -35.50, z-score = -2.24, R) >droEre2.scaffold_4784 13219085 95 - 25762168 AUGCCGCCGGUCAUGUCGGUUAUGGCCGUUGCUGUACUGUGGUCGCUGUCAAUGAAGGUCCAUAUGUCAAGGGACUUAGCAGCAGCAACGGCUCA .....((((((((((.....)))))))(((((((..((((((((.(((.(((((......))).)).).)).))))..))))))))))))))... ( -37.50, z-score = -2.02, R) >droAna3.scaffold_13337 11021423 90 + 23293914 --GCCGCCGGUUAUAUUGGUUAUGGCCGGUGCUGUACUGCGGUGUCCGUCAAUGCAAGUCCAUAUGUCAGGCGACUUAACAGCAACAACGGC--- --(((((((((((((.....)))))))))((((((...(((.((.....)).)))((((((.........).))))).))))))....))))--- ( -30.80, z-score = -0.70, R) >consensus AUGCCGCCGGUCAUGUCGGUUAUGGCCGUUGCUGUACUGUGGUCGUUGUCAAUGAAAGUCCAUAUGUCAAGGGACUUAGCAGCAGCAACGGCUCA ..((((..(((((((.....)))))))((((((((.....((((.(((.(((((......))).)).).)).))))..))))))))..))))... (-33.67 = -32.98 + -0.69)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:23:06 2011