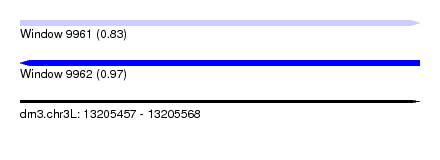
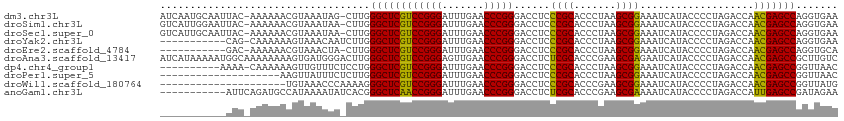
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 13,205,457 – 13,205,568 |
| Length | 111 |
| Max. P | 0.965166 |

| Location | 13,205,457 – 13,205,568 |
|---|---|
| Length | 111 |
| Sequences | 10 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 78.33 |
| Shannon entropy | 0.45480 |
| G+C content | 0.52830 |
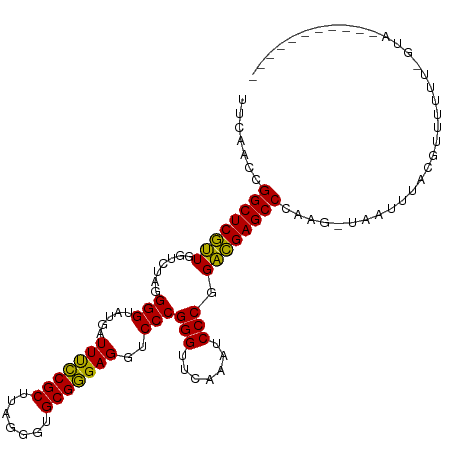
| Mean single sequence MFE | -34.77 |
| Consensus MFE | -33.07 |
| Energy contribution | -32.30 |
| Covariance contribution | -0.77 |
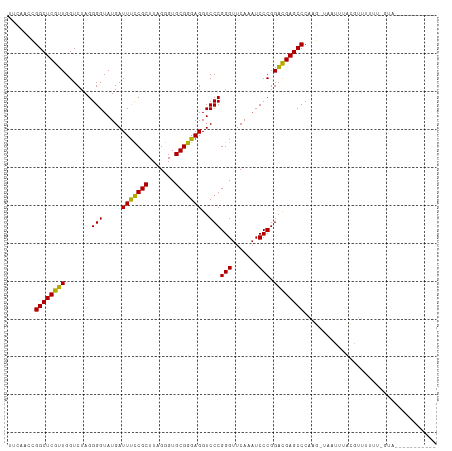
| Combinations/Pair | 1.19 |
| Mean z-score | -0.63 |
| Structure conservation index | 0.95 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.830149 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
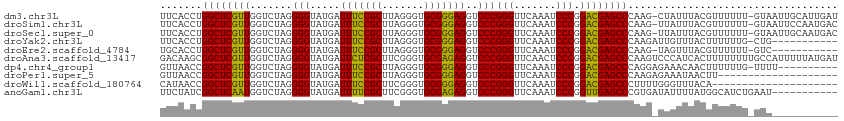
>dm3.chr3L 13205457 111 + 24543557 UUCACCUGGCUCGUUGGUCUAGGGGUAUGAUUUCCGCUUAGGGUGCGGGAGGUCCCGGGUUCAAAUCCCGGACGAGCCCAAG-CUAUUUACGUUUUUU-GUAAUUGCAUUGAU .......((((((((.......(((.....(((((((.......)))))))..)))(((.......))).))))))))((((-(...(((((.....)-))))..)).))).. ( -38.20, z-score = -1.28, R) >droSim1.chr3L 12600801 111 + 22553184 UUCACCUGGCUCGUUGGUCUAGGGGUAUGAUUUCCGCUUAGGGUGCGGGAGGUCCCGGGUUCAAAUCCCGGACGAGCCCAAG-UUAUUUACGUUUUUU-GUAAUUCCAAUGAC ..........(((((((.....((((....(((((((.......)))))))((((.(((.......)))))))..))))...-....(((((.....)-))))..))))))). ( -35.60, z-score = -0.71, R) >droSec1.super_0 5381067 111 + 21120651 UUCACCUGGCUCGUUGGUCUAGGGGUAUGAUUUCCGCUUAGGGUGCGGGAGGUCCCGGGUUCAAAUCCCGGACGAGCCCAAG-UUAUUUACGUUUUUU-GUAAUUGCAAUGAC .(((...((((((((.......(((.....(((((((.......)))))))..)))(((.......))).))))))))...(-(...(((((.....)-))))..))..))). ( -36.20, z-score = -0.94, R) >droYak2.chr3L 13292622 101 + 24197627 UUCACCUGGCUCGUUGGUCUAGGGGUAUGAUUUCCGCUUAGGGUGCGGGAGGUCCCGGGUUCAAAUCCCGGACGAGCCCAAGAUUGUUUACUUUUUUG-CUG----------- .......((((((((.......(((.....(((((((.......)))))))..)))(((.......))).))))))))(((((..(....)..)))))-...----------- ( -34.20, z-score = -0.55, R) >droEre2.scaffold_4784 13215390 100 + 25762168 UGCACCUGGCUCGUUGGUCUAGGGGUAUGAUUUCCGCUUAGGGUGCGGGAGGUCCCGGGUUCAAAUCCCGGACGAGCCCAAG-UAGUUUACGUUUUUU-GUC----------- (((....((((((((.......(((.....(((((((.......)))))))..)))(((.......))).))))))))...)-)).............-...----------- ( -32.70, z-score = -0.13, R) >droAna3.scaffold_13417 5088146 113 - 6960332 GACAAGCGGCUCGUUGGUCUAGGGGUAUGAUUCUCGCUUCGGGUGCGAGAGGUCCCGGGUUCAACUCCCGGACGAGCCCAAGUCCCAUCACUUUUUUUUGCCAUUUUUAUGAU (((..(.((((((((.......(((.....(((((((.......)))))))..)))(((.......))).)))))))))..)))............................. ( -36.00, z-score = -0.46, R) >dp4.chr4_group1 2040291 102 - 5278887 GUUAACCGGCUCGUUGGUCUAGGGGUAUGAUUUCCGCUUAGGGUGCGGGAGGUCCCGGGUUCAAAUCCCGGACGAGCCCAGGAGAAACAACUUUUUUG-UUUU---------- .....((((((((((.......(((.....(((((((.......)))))))..)))(((.......))).))))))))..)).(((((((.....)))-))))---------- ( -36.40, z-score = -0.94, R) >droPer1.super_5 1983899 93 + 6813705 GUUAACCGGCUCGUUGGUCUAGGGGUAUGAUUUCCGCUUAGGGUGCGGGAGGUCCCGGGUUCAAAUCCCGGACGAGCCCAAGAGAAAUAACUU-------------------- .......((((((((.......(((.....(((((((.......)))))))..)))(((.......))).))))))))...............-------------------- ( -32.70, z-score = -0.59, R) >droWil1.scaffold_180764 990239 92 - 3949147 CAUAACCGGCUCGUUGGUCUAGGGGUAUGAUUUCCGCUUCGGGUGCGGGAGGUCCCGGGUUCAAAUCCCGGACGAGCCCUUUUGGGUUUACA--------------------- ...((((((((((((.......(((.....(((((((.......)))))))..)))(((.......))).))))))))(....)))))....--------------------- ( -34.00, z-score = -0.37, R) >anoGam1.chr3L 28727965 102 - 41284009 UUCUAUCGGCUCAAUGGUCUAGGGGUAUGAUUUUCGCUUCGGGUGCGAGAGGUCCCGGGUUCAAAUCCCGGUUGAGCCCGUGAUAUUUUAUGGCAUCUGAAU----------- .....((((((((((.......(((.....(((((((.......)))))))..)))(((.......))).))))))))(((((....)))))......))..----------- ( -31.70, z-score = -0.31, R) >consensus UUCAACCGGCUCGUUGGUCUAGGGGUAUGAUUUCCGCUUAGGGUGCGGGAGGUCCCGGGUUCAAAUCCCGGACGAGCCCAAG_UAAUUUACGUUUUUU_GUA___________ .......((((((((.......(((.....(((((((.......)))))))..)))(((.......))).))))))))................................... (-33.07 = -32.30 + -0.77)
| Location | 13,205,457 – 13,205,568 |
|---|---|
| Length | 111 |
| Sequences | 10 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 78.33 |
| Shannon entropy | 0.45480 |
| G+C content | 0.52830 |
| Mean single sequence MFE | -29.94 |
| Consensus MFE | -28.02 |
| Energy contribution | -27.34 |
| Covariance contribution | -0.68 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.94 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.75 |
| SVM RNA-class probability | 0.965166 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
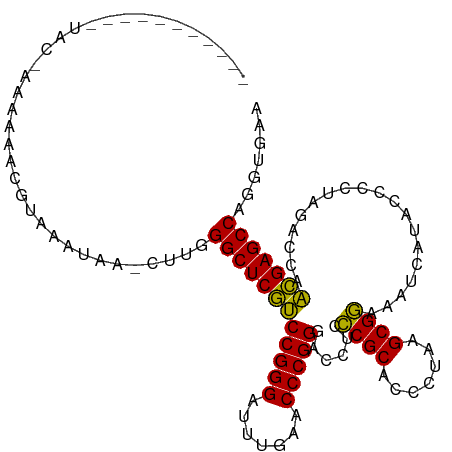
>dm3.chr3L 13205457 111 - 24543557 AUCAAUGCAAUUAC-AAAAAACGUAAAUAG-CUUGGGCUCGUCCGGGAUUUGAACCCGGGACCUCCCGCACCCUAAGCGGAAAUCAUACCCCUAGACCAACGAGCCAGGUGAA ..(((.((..((((-.......))))...)-))))((((((((((((.......)))))......((((.......))))...................)))))))....... ( -30.90, z-score = -1.36, R) >droSim1.chr3L 12600801 111 - 22553184 GUCAUUGGAAUUAC-AAAAAACGUAAAUAA-CUUGGGCUCGUCCGGGAUUUGAACCCGGGACCUCCCGCACCCUAAGCGGAAAUCAUACCCCUAGACCAACGAGCCAGGUGAA .(((((....((((-.......))))....-....((((((((((((.......)))))......((((.......))))...................))))))).))))). ( -31.20, z-score = -1.10, R) >droSec1.super_0 5381067 111 - 21120651 GUCAUUGCAAUUAC-AAAAAACGUAAAUAA-CUUGGGCUCGUCCGGGAUUUGAACCCGGGACCUCCCGCACCCUAAGCGGAAAUCAUACCCCUAGACCAACGAGCCAGGUGAA .(((((....((((-.......))))....-....((((((((((((.......)))))......((((.......))))...................))))))).))))). ( -31.20, z-score = -1.56, R) >droYak2.chr3L 13292622 101 - 24197627 -----------CAG-CAAAAAAGUAAACAAUCUUGGGCUCGUCCGGGAUUUGAACCCGGGACCUCCCGCACCCUAAGCGGAAAUCAUACCCCUAGACCAACGAGCCAGGUGAA -----------((.-(....(((........))).((((((((((((.......)))))......((((.......))))...................))))))).).)).. ( -28.70, z-score = -1.07, R) >droEre2.scaffold_4784 13215390 100 - 25762168 -----------GAC-AAAAAACGUAAACUA-CUUGGGCUCGUCCGGGAUUUGAACCCGGGACCUCCCGCACCCUAAGCGGAAAUCAUACCCCUAGACCAACGAGCCAGGUGCA -----------...-.............((-(((((.((((((((((.......)))))......((((.......))))...................)))))))))))).. ( -30.40, z-score = -1.71, R) >droAna3.scaffold_13417 5088146 113 + 6960332 AUCAUAAAAAUGGCAAAAAAAAGUGAUGGGACUUGGGCUCGUCCGGGAGUUGAACCCGGGACCUCUCGCACCCGAAGCGAGAAUCAUACCCCUAGACCAACGAGCCGCUUGUC ...........(((((....((((......)))).((((((((((((.......)))))((..((((((.......)))))).))..............)))))))..))))) ( -34.70, z-score = -1.18, R) >dp4.chr4_group1 2040291 102 + 5278887 ----------AAAA-CAAAAAAGUUGUUUCUCCUGGGCUCGUCCGGGAUUUGAACCCGGGACCUCCCGCACCCUAAGCGGAAAUCAUACCCCUAGACCAACGAGCCGGUUAAC ----------.(((-(((.....))))))......((((((((((((.......)))))......((((.......))))...................)))))))....... ( -29.40, z-score = -1.10, R) >droPer1.super_5 1983899 93 - 6813705 --------------------AAGUUAUUUCUCUUGGGCUCGUCCGGGAUUUGAACCCGGGACCUCCCGCACCCUAAGCGGAAAUCAUACCCCUAGACCAACGAGCCGGUUAAC --------------------...............((((((((((((.......)))))......((((.......))))...................)))))))....... ( -27.80, z-score = -0.84, R) >droWil1.scaffold_180764 990239 92 + 3949147 ---------------------UGUAAACCCAAAAGGGCUCGUCCGGGAUUUGAACCCGGGACCUCCCGCACCCGAAGCGGAAAUCAUACCCCUAGACCAACGAGCCGGUUAUG ---------------------....(((((....)((((((((((((.......)))))......((((.......))))...................)))))))))))... ( -28.70, z-score = -1.21, R) >anoGam1.chr3L 28727965 102 + 41284009 -----------AUUCAGAUGCCAUAAAAUAUCACGGGCUCAACCGGGAUUUGAACCCGGGACCUCUCGCACCCGAAGCGAAAAUCAUACCCCUAGACCAUUGAGCCGAUAGAA -----------.(((.((((........))))...((((((((((((.......)))))......((((.......))))...................)))))))....))) ( -26.40, z-score = -1.68, R) >consensus ___________UAC_AAAAAACGUAAAUAA_CUUGGGCUCGUCCGGGAUUUGAACCCGGGACCUCCCGCACCCUAAGCGGAAAUCAUACCCCUAGACCAACGAGCCAGGUGAA ...................................((((((((((((.......)))))......((((.......))))...................)))))))....... (-28.02 = -27.34 + -0.68)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:23:04 2011