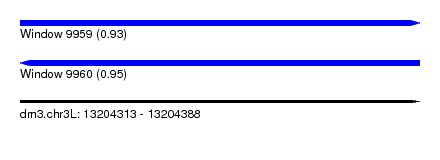
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 13,204,313 – 13,204,388 |
| Length | 75 |
| Max. P | 0.954383 |

| Location | 13,204,313 – 13,204,388 |
|---|---|
| Length | 75 |
| Sequences | 14 |
| Columns | 75 |
| Reading direction | forward |
| Mean pairwise identity | 91.94 |
| Shannon entropy | 0.21008 |
| G+C content | 0.61006 |
| Mean single sequence MFE | -31.61 |
| Consensus MFE | -29.26 |
| Energy contribution | -29.01 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.93 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.32 |
| SVM RNA-class probability | 0.925967 |
| Prediction | RNA |
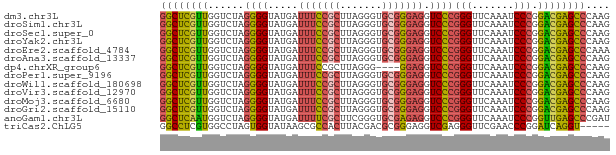
Download alignment: ClustalW | MAF
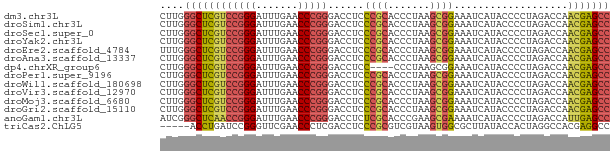
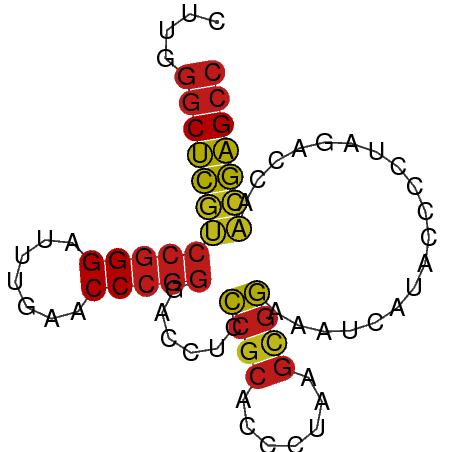
>dm3.chr3L 13204313 75 + 24543557 GGCUCGUUGGUCUAGGGGUAUGAUUUCCGCUUAGGGUGCGGGAGGUCCCGGGUUCAAAUCCCGGACGAGCCCAAG ((((((((.......(((.....(((((((.......)))))))..)))(((.......))).)))))))).... ( -32.40, z-score = -1.73, R) >droSim1.chr3L 12599696 75 + 22553184 GGCUCGUUGGUCUAGGGGUAUGAUUUCCGCUUAGGGUGCGGGAGGUCCCGGGUUCAAAUCCCGGACGAGCCCAAG ((((((((.......(((.....(((((((.......)))))))..)))(((.......))).)))))))).... ( -32.40, z-score = -1.73, R) >droSec1.super_0 5379960 75 + 21120651 GGCUCGUUGGUCUAGGGGUAUGAUUUCCGCUUAGGGUGCGGGAGGUCCCGGGUUCAAAUCCCGGACGAGCCCAAG ((((((((.......(((.....(((((((.......)))))))..)))(((.......))).)))))))).... ( -32.40, z-score = -1.73, R) >droYak2.chr3L 13291524 75 + 24197627 GGCUCGUUGGUCUAGGGGUAUGAUUUCCGCUUAGGGUGCGGGAGGUCCCGGGUUCAAAUCCCGGACGAGCCCAAG ((((((((.......(((.....(((((((.......)))))))..)))(((.......))).)))))))).... ( -32.40, z-score = -1.73, R) >droEre2.scaffold_4784 13214241 75 + 25762168 GGCUCGUUGGUCUAGGGGUAUGAUUUCCGCUUAGGGUGCGGGAGGUCCCGGGUUCAAAUCCCGGACGAGCCCAAA ((((((((.......(((.....(((((((.......)))))))..)))(((.......))).)))))))).... ( -32.40, z-score = -1.63, R) >droAna3.scaffold_13337 11017576 75 - 23293914 GGCUCGUUGGUCUAGGGGUAUGAUUUCCGCUUAGGGUGCGGGAGGUCCCGGGUUCAAAUCCCGGACGAGCCCAAG ((((((((.......(((.....(((((((.......)))))))..)))(((.......))).)))))))).... ( -32.40, z-score = -1.73, R) >dp4.chrXR_group6 7897695 71 - 13314419 GGCUCGUUGGUCUAGGGGUAUGAUUUCCGCUUAGGG----GGAGGUCCCGGGUUCAAAUCCCGGACGAGCCCAAG (((((((..............(((((((.(....).----)))))))(((((.......)))))))))))).... ( -30.60, z-score = -1.61, R) >droPer1.super_9196 809 75 + 1238 GGCUCGUUGGUCUAGGGGUAUGAUUUCCGCUUAGGGUGCGGGAGGUCCCGGGUUCAAAUCCCGGACGAGCCCAAG ((((((((.......(((.....(((((((.......)))))))..)))(((.......))).)))))))).... ( -32.40, z-score = -1.73, R) >droWil1.scaffold_180698 6735092 75 + 11422946 GGCUCGUUGGUCUAGGGGUAUGAUUUCCGCUUAGGGUGCGGGAGGUCCCGGGUUCAAAUCCCGGACGAGCCCAAG ((((((((.......(((.....(((((((.......)))))))..)))(((.......))).)))))))).... ( -32.40, z-score = -1.73, R) >droVir3.scaffold_12970 6178753 75 - 11907090 GGCUCGUUGGUCUAGGGGUAUGAUUUCCGCUUAGGGUGCGGGAGGUCCCGGGUUCAAAUCCCGGACGAGCCCAAG ((((((((.......(((.....(((((((.......)))))))..)))(((.......))).)))))))).... ( -32.40, z-score = -1.73, R) >droMoj3.scaffold_6680 9923560 75 - 24764193 GGCUCGUUGGUCUAGGGGUAUGAUUUCCGCUUAGGGUGCGGGAGGUCCCGGGUUCAAAUCCCGGACGAGCCCAAG ((((((((.......(((.....(((((((.......)))))))..)))(((.......))).)))))))).... ( -32.40, z-score = -1.73, R) >droGri2.scaffold_15110 23222284 75 + 24565398 GGCUCGUUGGUCUAGGGGUAUGAUUUCCGCUUAGGGUGCGGGAGGUCCCGGGUUCAAAUCCCGGACGAGCCCAAG ((((((((.......(((.....(((((((.......)))))))..)))(((.......))).)))))))).... ( -32.40, z-score = -1.73, R) >anoGam1.chr3L 28727336 75 - 41284009 GGCUCAAUGGUCUAGGGGUAUGAUUUUCGCUUCGGGUGCGAGAGGUCCCGGGUUCAAAUCCCGGUUGAGCCCGAU ((((((((.......(((.....(((((((.......)))))))..)))(((.......))).)))))))).... ( -29.10, z-score = -1.26, R) >triCas2.ChLG5 14942443 70 + 18847211 GGCCUCGUGGCCUAGUGGUAUAAGCGCCACUUACGACGCGGGAGGUCGAGGGUUCGAACCCGGAUCAGGU----- ...((((((....((((((......)))))).....)))))).((((..((((....)))).))))....----- ( -26.50, z-score = -0.60, R) >consensus GGCUCGUUGGUCUAGGGGUAUGAUUUCCGCUUAGGGUGCGGGAGGUCCCGGGUUCAAAUCCCGGACGAGCCCAAG ((((((((......((((.....(((((((.......))))))).))))(((.......))).)))))))).... (-29.26 = -29.01 + -0.25)
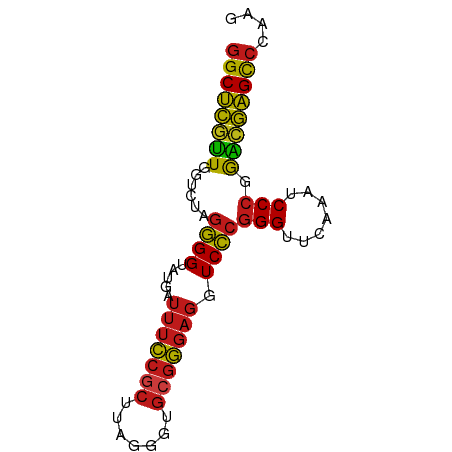
| Location | 13,204,313 – 13,204,388 |
|---|---|
| Length | 75 |
| Sequences | 14 |
| Columns | 75 |
| Reading direction | reverse |
| Mean pairwise identity | 91.94 |
| Shannon entropy | 0.21008 |
| G+C content | 0.61006 |
| Mean single sequence MFE | -25.56 |
| Consensus MFE | -23.51 |
| Energy contribution | -23.52 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.92 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.60 |
| SVM RNA-class probability | 0.954383 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 13204313 75 - 24543557 CUUGGGCUCGUCCGGGAUUUGAACCCGGGACCUCCCGCACCCUAAGCGGAAAUCAUACCCCUAGACCAACGAGCC ....((((((((((((.......)))))......((((.......))))...................))))))) ( -26.10, z-score = -1.94, R) >droSim1.chr3L 12599696 75 - 22553184 CUUGGGCUCGUCCGGGAUUUGAACCCGGGACCUCCCGCACCCUAAGCGGAAAUCAUACCCCUAGACCAACGAGCC ....((((((((((((.......)))))......((((.......))))...................))))))) ( -26.10, z-score = -1.94, R) >droSec1.super_0 5379960 75 - 21120651 CUUGGGCUCGUCCGGGAUUUGAACCCGGGACCUCCCGCACCCUAAGCGGAAAUCAUACCCCUAGACCAACGAGCC ....((((((((((((.......)))))......((((.......))))...................))))))) ( -26.10, z-score = -1.94, R) >droYak2.chr3L 13291524 75 - 24197627 CUUGGGCUCGUCCGGGAUUUGAACCCGGGACCUCCCGCACCCUAAGCGGAAAUCAUACCCCUAGACCAACGAGCC ....((((((((((((.......)))))......((((.......))))...................))))))) ( -26.10, z-score = -1.94, R) >droEre2.scaffold_4784 13214241 75 - 25762168 UUUGGGCUCGUCCGGGAUUUGAACCCGGGACCUCCCGCACCCUAAGCGGAAAUCAUACCCCUAGACCAACGAGCC ....((((((((((((.......)))))......((((.......))))...................))))))) ( -26.10, z-score = -1.93, R) >droAna3.scaffold_13337 11017576 75 + 23293914 CUUGGGCUCGUCCGGGAUUUGAACCCGGGACCUCCCGCACCCUAAGCGGAAAUCAUACCCCUAGACCAACGAGCC ....((((((((((((.......)))))......((((.......))))...................))))))) ( -26.10, z-score = -1.94, R) >dp4.chrXR_group6 7897695 71 + 13314419 CUUGGGCUCGUCCGGGAUUUGAACCCGGGACCUCC----CCCUAAGCGGAAAUCAUACCCCUAGACCAACGAGCC ....((((((((((((.......)))))((..(((----........)))..))..............))))))) ( -22.70, z-score = -0.97, R) >droPer1.super_9196 809 75 - 1238 CUUGGGCUCGUCCGGGAUUUGAACCCGGGACCUCCCGCACCCUAAGCGGAAAUCAUACCCCUAGACCAACGAGCC ....((((((((((((.......)))))......((((.......))))...................))))))) ( -26.10, z-score = -1.94, R) >droWil1.scaffold_180698 6735092 75 - 11422946 CUUGGGCUCGUCCGGGAUUUGAACCCGGGACCUCCCGCACCCUAAGCGGAAAUCAUACCCCUAGACCAACGAGCC ....((((((((((((.......)))))......((((.......))))...................))))))) ( -26.10, z-score = -1.94, R) >droVir3.scaffold_12970 6178753 75 + 11907090 CUUGGGCUCGUCCGGGAUUUGAACCCGGGACCUCCCGCACCCUAAGCGGAAAUCAUACCCCUAGACCAACGAGCC ....((((((((((((.......)))))......((((.......))))...................))))))) ( -26.10, z-score = -1.94, R) >droMoj3.scaffold_6680 9923560 75 + 24764193 CUUGGGCUCGUCCGGGAUUUGAACCCGGGACCUCCCGCACCCUAAGCGGAAAUCAUACCCCUAGACCAACGAGCC ....((((((((((((.......)))))......((((.......))))...................))))))) ( -26.10, z-score = -1.94, R) >droGri2.scaffold_15110 23222284 75 - 24565398 CUUGGGCUCGUCCGGGAUUUGAACCCGGGACCUCCCGCACCCUAAGCGGAAAUCAUACCCCUAGACCAACGAGCC ....((((((((((((.......)))))......((((.......))))...................))))))) ( -26.10, z-score = -1.94, R) >anoGam1.chr3L 28727336 75 + 41284009 AUCGGGCUCAACCGGGAUUUGAACCCGGGACCUCUCGCACCCGAAGCGAAAAUCAUACCCCUAGACCAUUGAGCC ....((((((((((((.......)))))......((((.......))))...................))))))) ( -23.40, z-score = -1.91, R) >triCas2.ChLG5 14942443 70 - 18847211 -----ACCUGAUCCGGGUUCGAACCCUCGACCUCCCGCGUCGUAAGUGGCGCUUAUACCACUAGGCCACGAGGCC -----....(((.((((.((((....))))...)))).)))...(((((........))))).((((....)))) ( -24.60, z-score = -1.54, R) >consensus CUUGGGCUCGUCCGGGAUUUGAACCCGGGACCUCCCGCACCCUAAGCGGAAAUCAUACCCCUAGACCAACGAGCC ....((((((((((((.......)))))......((((.......))))...................))))))) (-23.51 = -23.52 + 0.01)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:23:02 2011