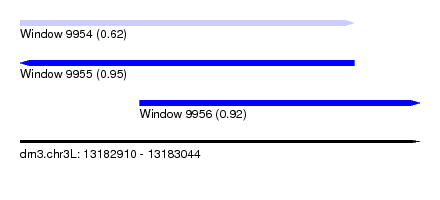
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 13,182,910 – 13,183,044 |
| Length | 134 |
| Max. P | 0.953491 |

| Location | 13,182,910 – 13,183,022 |
|---|---|
| Length | 112 |
| Sequences | 10 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 78.61 |
| Shannon entropy | 0.42256 |
| G+C content | 0.47345 |
| Mean single sequence MFE | -26.15 |
| Consensus MFE | -14.86 |
| Energy contribution | -14.97 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.620029 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
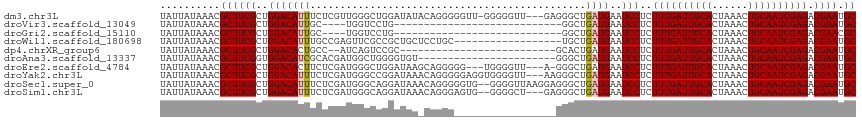
>dm3.chr3L 13182910 112 + 24543557 UAUUAUAAACGCUUCGCUGGACAUUUCUCGUUGGGCUGGAUAUACAGGGGGUU-GGGGGUU---GAGGGCUGAUGAAUCCUCUUUGAUUGCACUAAACUGCAAUCGAGACGAAUGC ..........((((((((.(((.......))).))).........((((((((-.(.(((.---....)))..).))))))))(((((((((......)))))))))...))).)) ( -29.20, z-score = -0.93, R) >droVir3.scaffold_13049 12570583 84 + 25233164 UAUUAUAAACGCUUCGCUGGACAUUGC----UGGUCCUG----------------------------GGCUGAUGAAUCCUCUUUGAUUGCACUAAACUGCAAUCGAGACGAAUGC ..........((((((((((((.....----..))))..----------------------------)))............((((((((((......))))))))))..))).)) ( -20.30, z-score = -1.83, R) >droGri2.scaffold_15110 1420884 84 - 24565398 UAUUAUAAACGCUUCGCUGGACAUUGC----UGGUCCUG----------------------------GGCUGAUGAAUCCUCUUUGAUUGCACUAAACUGCAAUCGAGACGAACGC ..........((((((((((((.....----..))))..----------------------------)))............((((((((((......))))))))))..))).)) ( -20.30, z-score = -1.83, R) >droWil1.scaffold_180698 4719508 98 - 11422946 UAUUAUAAACGCUUCGCUGGACAUUUGCCGAGUUCGCCGCUGCUCCUGC------------------UGCUGAUGAAUCCUCUUUGAUUGCACUAAACUGCAAUCGAGACGAAUGC ..........((((((..((.(....)))((((((((.((.((....))------------------.)).).))))..)))((((((((((......)))))))))).)))).)) ( -23.40, z-score = -1.43, R) >dp4.chrXR_group6 5445136 88 + 13314419 UAUUAUAAACGCUUCGCUGGACACUGCC--AUCAGUCCGC--------------------------GCACUGAUGAAUCCUCUUUGAUUGCACUAAACUGCAAUCGAGACGAAUGC ..........((((((..(((......(--((((((....--------------------------..)))))))..)))..((((((((((......)))))))))).)))).)) ( -23.10, z-score = -2.76, R) >droAna3.scaffold_13337 10997262 93 - 23293914 UAUUAUAAACGCUUCGGUGGACAUCGCACGAUGGCUGGGGUGU-----------------------GGGCUGAUGAAUCCUCUUUGAUUGCACUAAACUGCAAUCGAGACGAAUGC (((((...((((((((((...((((....))))))))))))))-----------------------....)))))..((.((((.(((((((......))))))))))).)).... ( -28.30, z-score = -2.39, R) >droEre2.scaffold_4784 13192790 109 + 25762168 UAUUAUAAACGCUUCGCUGGACACUUCUCGAUGGGCUGGAUAAGCAGGGGG---UGGGGUU---A-GGGCUGAUGAAUCCUCUUUGAUUGCACUAAACUGCAAUCGAGACGAAUGC ..........((((((((.((((.(((.....))).)).....((((..((---((.((((---(-(((..((....))..)))))))).))))...))))..)).)).)))).)) ( -28.50, z-score = -0.76, R) >droYak2.chr3L 13269521 113 + 24197627 UAUUAUAAACGCUUCGCUGGACAUUUCUCGAUGGGCCGGAUAAACAGGGGGAGGUGGGGUU---AAGGGCUGAUGAAUCCUCUUUGAUUGCACUAAACUGCAAUCGAGACGAAUGC ..........((((((((((.((((....))))..)))).......((((((.((.((.(.---...).)).))...))))))(((((((((......)))))))))..)))).)) ( -28.50, z-score = -0.52, R) >droSec1.super_0 5358575 114 + 21120651 UAUUAUAAACGCUUCGCUGGACAUUUCUCGAUGGGCAGGAUAAACAGGGGGUG--GGGGUUAAGGAGGGCUGAUGAAUCCUCUUUGAUUGCACUAAACUGCAAUCGAGACGAAUGC ..........((((((.........(((((((..((((...........((((--.((.((((((((((........)))))))))))).))))...)))).))))))))))).)) ( -32.44, z-score = -2.03, R) >droSim1.chr3L 12578312 111 + 22553184 UAUUAUAAACGCUUCGCUGGACAUUUCUCGAUGGGCAGGAUAAACAGGGAGUG--GGGGCU---GAGGGCUGAUGAAUCCUCUUUGAUUGCACUAAACUGCAAUCGAGACGAAUGC .........((((((.(((.....(((((.....).))))....)))))))))--......---(((((........)))))((((((((((......))))))))))........ ( -27.50, z-score = -0.55, R) >consensus UAUUAUAAACGCUUCGCUGGACAUUUCUCGAUGGGCCGGAUAA_CAGGG_________________GGGCUGAUGAAUCCUCUUUGAUUGCACUAAACUGCAAUCGAGACGAAUGC ..........((((((..(((((((..............................................))))..)))..((((((((((......)))))))))).)))).)) (-14.86 = -14.97 + 0.11)
| Location | 13,182,910 – 13,183,022 |
|---|---|
| Length | 112 |
| Sequences | 10 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 78.61 |
| Shannon entropy | 0.42256 |
| G+C content | 0.47345 |
| Mean single sequence MFE | -22.81 |
| Consensus MFE | -16.79 |
| Energy contribution | -16.89 |
| Covariance contribution | 0.10 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.953491 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 13182910 112 - 24543557 GCAUUCGUCUCGAUUGCAGUUUAGUGCAAUCAAAGAGGAUUCAUCAGCCCUC---AACCCCC-AACCCCCUGUAUAUCCAGCCCAACGAGAAAUGUCCAGCGAAGCGUUUAUAAUA ((.(((((((((((((((......)))))))...((((..........))))---.......-......(((......)))......))).........))))))).......... ( -21.80, z-score = -2.27, R) >droVir3.scaffold_13049 12570583 84 - 25233164 GCAUUCGUCUCGAUUGCAGUUUAGUGCAAUCAAAGAGGAUUCAUCAGCC----------------------------CAGGACCA----GCAAUGUCCAGCGAAGCGUUUAUAAUA ((.(((((((((((((((......)))))))...)))............----------------------------..((((.(----....))))).))))))).......... ( -21.50, z-score = -2.23, R) >droGri2.scaffold_15110 1420884 84 + 24565398 GCGUUCGUCUCGAUUGCAGUUUAGUGCAAUCAAAGAGGAUUCAUCAGCC----------------------------CAGGACCA----GCAAUGUCCAGCGAAGCGUUUAUAAUA ((.(((((((((((((((......)))))))...)))............----------------------------..((((.(----....))))).))))))).......... ( -21.50, z-score = -1.92, R) >droWil1.scaffold_180698 4719508 98 + 11422946 GCAUUCGUCUCGAUUGCAGUUUAGUGCAAUCAAAGAGGAUUCAUCAGCA------------------GCAGGAGCAGCGGCGAACUCGGCAAAUGUCCAGCGAAGCGUUUAUAAUA ((.(((((...(((((((......))))))).....((((......((.------------------((....)).))(.((....)).)....)))).))))))).......... ( -26.40, z-score = -0.72, R) >dp4.chrXR_group6 5445136 88 - 13314419 GCAUUCGUCUCGAUUGCAGUUUAGUGCAAUCAAAGAGGAUUCAUCAGUGC--------------------------GCGGACUGAU--GGCAGUGUCCAGCGAAGCGUUUAUAAUA ((.(((((...(((((((......))))))).....((((((((((((..--------------------------....))))))--))....)))).))))))).......... ( -28.90, z-score = -2.24, R) >droAna3.scaffold_13337 10997262 93 + 23293914 GCAUUCGUCUCGAUUGCAGUUUAGUGCAAUCAAAGAGGAUUCAUCAGCCC-----------------------ACACCCCAGCCAUCGUGCGAUGUCCACCGAAGCGUUUAUAAUA ((.((((....(((((((......))))))).....(((..((((.((.(-----------------------..............).)))))))))..)))))).......... ( -20.24, z-score = -1.48, R) >droEre2.scaffold_4784 13192790 109 - 25762168 GCAUUCGUCUCGAUUGCAGUUUAGUGCAAUCAAAGAGGAUUCAUCAGCCC-U---AACCCCA---CCCCCUGCUUAUCCAGCCCAUCGAGAAGUGUCCAGCGAAGCGUUUAUAAUA ((.(((.(((((((((((......))))))).....((((....(((...-.---.......---....)))...))))........)))).((.....))))))).......... ( -21.26, z-score = -1.41, R) >droYak2.chr3L 13269521 113 - 24197627 GCAUUCGUCUCGAUUGCAGUUUAGUGCAAUCAAAGAGGAUUCAUCAGCCCUU---AACCCCACCUCCCCCUGUUUAUCCGGCCCAUCGAGAAAUGUCCAGCGAAGCGUUUAUAAUA ((.(((((((((((((((......))))))).....((((....(((.....---..............)))...))))........))).........))))))).......... ( -21.41, z-score = -0.96, R) >droSec1.super_0 5358575 114 - 21120651 GCAUUCGUCUCGAUUGCAGUUUAGUGCAAUCAAAGAGGAUUCAUCAGCCCUCCUUAACCCC--CACCCCCUGUUUAUCCUGCCCAUCGAGAAAUGUCCAGCGAAGCGUUUAUAAUA ((.(((((((((((((((......)))))))....(((((....(((..............--......)))...))))).......))).........))))))).......... ( -22.15, z-score = -1.94, R) >droSim1.chr3L 12578312 111 - 22553184 GCAUUCGUCUCGAUUGCAGUUUAGUGCAAUCAAAGAGGAUUCAUCAGCCCUC---AGCCCC--CACUCCCUGUUUAUCCUGCCCAUCGAGAAAUGUCCAGCGAAGCGUUUAUAAUA ((.(((((((((((.((((..(((.(((......((((..........))))---((....--..))...))))))..))))..)))))).........))))))).......... ( -22.90, z-score = -1.46, R) >consensus GCAUUCGUCUCGAUUGCAGUUUAGUGCAAUCAAAGAGGAUUCAUCAGCCC_________________CCCUG_UUAUCCGGCCCAUCGAGAAAUGUCCAGCGAAGCGUUUAUAAUA ((.(((((...(((((((......))))))).....((((......................................................)))).))))))).......... (-16.79 = -16.89 + 0.10)
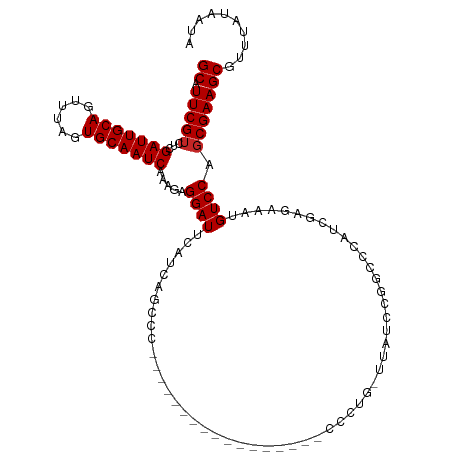
| Location | 13,182,950 – 13,183,044 |
|---|---|
| Length | 94 |
| Sequences | 11 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 76.03 |
| Shannon entropy | 0.43546 |
| G+C content | 0.49165 |
| Mean single sequence MFE | -24.99 |
| Consensus MFE | -16.89 |
| Energy contribution | -16.58 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.917276 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 13182950 94 + 24543557 UAUACAGGGGGUU-GGGGGUUGA---GGGCUGAUGAAUCCUCUUUGAUUGCACUAAACUGCAAUCGAGACGAAUGCCCAAAACGUGCGACGUCAUGUG------------------ ..((((.((.(((-(.(.(((..---((((.......((.((((.(((((((......))))))))))).))..))))..))).).)))).)).))))------------------ ( -32.10, z-score = -2.55, R) >droVir3.scaffold_13049 12570611 78 + 25233164 ------------------GGUCCUGG--GCUGAUGAAUCCUCUUUGAUUGCACUAAACUGCAAUCGAGACGAAUGCGCAAAACGUGCAAUGUCAUGCG------------------ ------------------........--((((((...((.((((.(((((((......))))))))))).)).(((((.....)))))..)))).)).------------------ ( -23.10, z-score = -2.11, R) >droMoj3.scaffold_6680 9895760 80 - 24764193 ------------------GGUCCUGGGCUCUGAUGAAUCCUCUUUGAUUGCACUAAACUGCAAUCGAGACGAAUGCGCAAAACGUGCAAUGUCAUGCG------------------ ------------------.(((..((..((....))..))...(((((((((......))))))))))))((.(((((.....)))))...)).....------------------ ( -23.50, z-score = -2.21, R) >droGri2.scaffold_15110 1420912 78 - 24565398 ------------------GGUCCUGG--GCUGAUGAAUCCUCUUUGAUUGCACUAAACUGCAAUCGAGACGAACGCGCAAAACGUGCAAUGUCAUGCG------------------ ------------------........--((((((...((.((((.(((((((......))))))))))).))..((((.....))))...)))).)).------------------ ( -22.40, z-score = -1.80, R) >droWil1.scaffold_180698 4719540 106 - 11422946 ---------UUCGCCGCUGCUCCUGCU-GCUGAUGAAUCCUCUUUGAUUGCACUAAACUGCAAUCGAGACGAAUGCUCAAAACGUGCAAUGUCAUGCGUCCUUCGUACUCAAAAAA ---------(((((.((.((....)).-)).).))))........(((((((......)))))))((((((((.((.((..((((...))))..)).))..))))).)))...... ( -24.80, z-score = -2.19, R) >dp4.chrXR_group6 5445163 85 + 13314419 --------------CAUCAGUCC-GCGCACUGAUGAAUCCUCUUUGAUUGCACUAAACUGCAAUCGAGACGAAUGCUCAAAACGUGCAAUGUCAUGCGGC---------------- --------------(((((((..-....)))))))...((...(((((((((......)))))))))((((..(((.(.....).))).))))....)).---------------- ( -24.20, z-score = -1.80, R) >droAna3.scaffold_13337 10997298 79 - 23293914 ---------------GGGGUGU----GGGCUGAUGAAUCCUCUUUGAUUGCACUAAACUGCAAUCGAGACGAAUGCCCAAAACGUGCGAUGUCAUCUC------------------ ---------------(((((((----((((.......((.((((.(((((((......))))))))))).))..)))))..((((...))))))))))------------------ ( -22.60, z-score = -1.61, R) >droEre2.scaffold_4784 13192830 91 + 25762168 UAAGCAGGGGG---UGGGGUUA----GGGCUGAUGAAUCCUCUUUGAUUGCACUAAACUGCAAUCGAGACGAAUGCCCAAAACGUGCGACGUCAUGUG------------------ ...(((.(...---((((((((----....))))...((.((((.(((((((......))))))))))).))...))))...).)))...........------------------ ( -25.50, z-score = -0.55, R) >droYak2.chr3L 13269561 95 + 24197627 UAAACAGGGGGAGGUGGGGUUAA---GGGCUGAUGAAUCCUCUUUGAUUGCACUAAACUGCAAUCGAGACGAAUGCCCAAAACGUGCGACGUCAUGUG------------------ ...(((....((.((.(.(((..---((((.......((.((((.(((((((......))))))))))).))..))))..)))...).)).)).))).------------------ ( -24.60, z-score = -0.38, R) >droSec1.super_0 5358615 96 + 21120651 UAAACAGGGGGUG--GGGGUUAAGGAGGGCUGAUGAAUCCUCUUUGAUUGCACUAAACUGCAAUCGAGACGAAUGCCUAAAACGUGCGACGUCAUGUG------------------ ...(((.((.((.--..((((((((((((........))))))))))))((((......(((.(((...))).))).......)))).)).)).))).------------------ ( -25.62, z-score = -0.93, R) >droSim1.chr3L 12578352 93 + 22553184 UAAACAGGGAGUG--GGGGCUGA---GGGCUGAUGAAUCCUCUUUGAUUGCACUAAACUGCAAUCGAGACGAAUGCCCAAAACGUGCGACGUCAUGUG------------------ .............--.((((.((---(((........)))))((((((((((......))))))))))......))))...(((((......))))).------------------ ( -26.50, z-score = -1.13, R) >consensus _______________GGGGUUCC_G_GGGCUGAUGAAUCCUCUUUGAUUGCACUAAACUGCAAUCGAGACGAAUGCCCAAAACGUGCGAUGUCAUGUG__________________ ............................(((((((..((.((((.(((((((......))))))))))).)).(((.(.....).))).))))).))................... (-16.89 = -16.58 + -0.31)

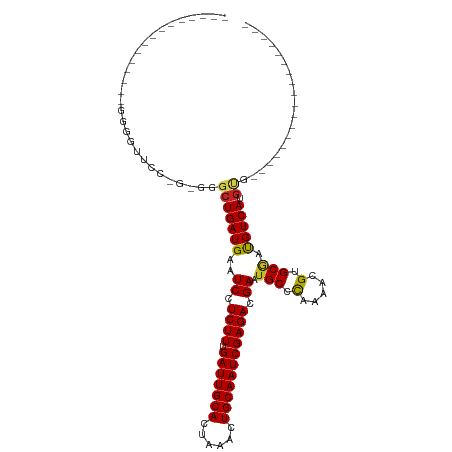
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:22:59 2011