| Sequence ID | dm3.chr3L |
|---|---|
| Location | 13,177,542 – 13,177,639 |
| Length | 97 |
| Max. P | 0.944825 |

| Location | 13,177,542 – 13,177,639 |
|---|---|
| Length | 97 |
| Sequences | 10 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 78.61 |
| Shannon entropy | 0.45206 |
| G+C content | 0.39915 |
| Mean single sequence MFE | -25.53 |
| Consensus MFE | -15.17 |
| Energy contribution | -14.57 |
| Covariance contribution | -0.60 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.944825 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
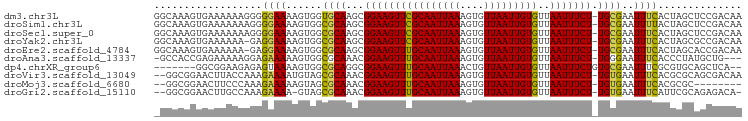
>dm3.chr3L 13177542 97 - 24543557 GGCAAAGUGAAAAAAGGGGGAAAAGUGGUGCAAGCGGAAGUUCGCAAUUAAAGUGUUAAUUGUGUUAAUUUCU-UGCGAAUUUCACUAGCUCCGACAA ................((((...(((((.....((((((((((((((((((....)))))))))..)))))).-))).....)))))..))))..... ( -25.00, z-score = -1.79, R) >droSim1.chr3L 12572981 97 - 22553184 GGCAAAGUGAAAAAAAGGGGAAAAGUGGCGCAAGCGGAAGUUCGCAAUUAAAGUGUUAAUUGUGUUAAUUUCU-UGCGAAUUUUACUAGCUCCGACAA ................((((...((((((....))((((((((((((((((....)))))))))..)))))))-.........))))..))))..... ( -23.50, z-score = -1.18, R) >droSec1.super_0 5353277 97 - 21120651 GGCAAAGUGAAAAAAAGGGGAAAAGUGGCGCAAGCGGAAGUUCGCAAUUAAAGUGUUAAUUGUGUUAAUUUCU-UGCGAAUUUCACUAGCUCCGACAA ................((((...((((((....))((((((((((((((((....)))))))))..)))))))-.........))))..))))..... ( -25.50, z-score = -1.72, R) >droYak2.chr3L 13264149 96 - 24197627 GGCAAAGUGAAAAAA-GAGGAAAAGUGGCGCAAGCGGAAGUUCGCAAUUAAAGUGUUAAUUGUGUUAAUUUCU-UGCGAAUUUCACUAGCGCCGACAA (((..(((((((...-...........((....))((((((((((((((((....)))))))))..)))))))-......)))))))...)))..... ( -27.40, z-score = -2.16, R) >droEre2.scaffold_4784 13186003 96 - 25762168 GGCAAAGUGAAAAAA-GAGGAAAAGUGGCGCAAGCGGAAGUUUGCAAUUAAAGUGUUAAUUGUGUUAAUUUCU-UGCGAAUUUCACUAGCACCGACAA .((..(((((((...-(((((((..((((((((((....))))))((((((....))))))..)))).)))))-).)...))))))).))........ ( -25.80, z-score = -2.07, R) >droAna3.scaffold_13337 10992044 93 + 23293914 -GCCACCGAGAAAAAGGAGAAAAAGUGGCGCAAACGGAAGUUUGCAAUUAAAGUGUUAAUUGUGUUAAUUUCU-UGGGAAUUUCACCCUAUGCUG--- -(((((..................)))))((((((....))))))......(((((.....(((..(((((..-...))))).)))...))))).--- ( -23.17, z-score = -1.55, R) >dp4.chrXR_group6 5439599 89 - 13314419 -------GGCGGAAGAGAGUAAAAGUGGCGCAGGCGGAAGUUUGCAAUUAAACUGUUAAUUGUGUUAAUUUCUGUGCGAAUUUCGCGUGCAGCUCA-- -------..(....).((((.......((((..(((((((((.((((((((....))))))))...)))))))))(((.....))))))).)))).-- ( -27.40, z-score = -1.58, R) >droVir3.scaffold_13049 12564662 95 - 25233164 --GGCGGAACUUACCAAAGAAAAUGUAGCGCAAACGGAAGUUUGCAAUUAAAGUGUUAAUUGUGUUAAUUUCU-UGUGAAUUUCACGCGCAGCGACAA --.((((((.((((..((((((...((((((((((....))))))((((((....))))))..)))).)))))-)))))..))).))).......... ( -24.90, z-score = -1.41, R) >droMoj3.scaffold_6680 9888652 87 + 24764193 --GGCGGAACUUCCCAAAGAAAAAGUAGCGCAAACGGAAGUUUGCAAUUAAAGUGUUAAUUGUGUUAAUUUCU-UGUGAAUUUCACGCGC-------- --.((((((.(((...((((((...((((((((((....))))))((((((....))))))..)))).)))))-)..))).))).)))..-------- ( -27.10, z-score = -3.39, R) >droGri2.scaffold_15110 1413958 93 + 24565398 --GGCGGAACUUGCCAAAGAAAA-GUAGCGCAAACGGAAGUUUGCAAUUAAAGUGUUAAUUGUGUUAAUUUCU-UGUGAAUUUCAUUCGCAGAGACA- --((((.....)))).((((((.-.((((((((((....))))))((((((....))))))..)))).)))))-)((((((...)))))).......- ( -25.50, z-score = -2.21, R) >consensus _GCAAAGUGAAAAAAAGAGGAAAAGUGGCGCAAGCGGAAGUUUGCAAUUAAAGUGUUAAUUGUGUUAAUUUCU_UGCGAAUUUCACUAGCACCGACAA ..................((((......((((...((((((((((((((((....)))))))))..))))))).))))..)))).............. (-15.17 = -14.57 + -0.60)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:22:57 2011