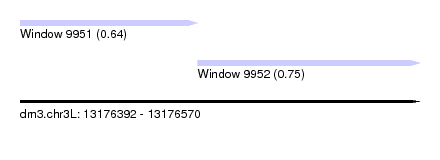
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 13,176,392 – 13,176,570 |
| Length | 178 |
| Max. P | 0.745822 |

| Location | 13,176,392 – 13,176,471 |
|---|---|
| Length | 79 |
| Sequences | 12 |
| Columns | 82 |
| Reading direction | forward |
| Mean pairwise identity | 79.50 |
| Shannon entropy | 0.42981 |
| G+C content | 0.41339 |
| Mean single sequence MFE | -15.98 |
| Consensus MFE | -8.27 |
| Energy contribution | -8.27 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.644950 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
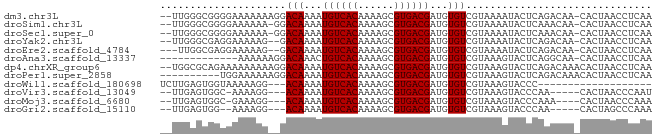
>dm3.chr3L 13176392 79 + 24543557 --UUGGGCGGGGAAAAAAAGGACAAAAUGUCACAAAAGCGUGACGAUGUGUCGUAAAAUACUCAGACAA-CACUAACCUCAA --......((((........((((...((((((......))))))...))))(((...)))........-......)))).. ( -17.20, z-score = -1.60, R) >droSim1.chr3L 12571827 78 + 22553184 --UUGGGCGGGGAAAAAA-GGACAAAAUGUCACAAAAGCGUGACGAUGUGUCGUAAAAUACUCAAACAA-CACUAACCUCAA --......((((......-.((((...((((((......))))))...))))(((...)))........-......)))).. ( -17.20, z-score = -1.87, R) >droSec1.super_0 5352151 78 + 21120651 --UUGGGCGGGGAAAAAA-GGACAAAAUGUCACAAAAGCGUGACGAUGUGUCGUAAAAUACUCAAACAA-CACUAACCUCAA --......((((......-.((((...((((((......))))))...))))(((...)))........-......)))).. ( -17.20, z-score = -1.87, R) >droYak2.chr3L 13263008 77 + 24197627 --UUGGGCGAGGAAAAAG--GACAAAAUGUCACAAAAGCGUGACGAUGUGUCGUAAAAUACUCAGACAA-CACUAACCUCAA --......((((......--((((...((((((......))))))...))))(((...)))........-......)))).. ( -18.10, z-score = -2.02, R) >droEre2.scaffold_4784 13184892 76 + 25762168 ---UUGGCGAGGAAAAAG--GACAAAAUGUCACAAAAGCGUGACGAUGUGUCGUAAAAUACUCAGACAA-CACUAACCUCAA ---.....((((......--((((...((((((......))))))...))))(((...)))........-......)))).. ( -18.10, z-score = -2.24, R) >droAna3.scaffold_13337 10991025 68 - 23293914 -------------AAAAAAGGACAAACUGUCACAAAAGCGUGACGAUGUGUCGUAAAGUACUCAGGCAA-CACUAACCUCAA -------------.......((((...((((((......))))))...))))............(....-)........... ( -13.70, z-score = -1.20, R) >dp4.chrXR_group6 5437614 80 + 13314419 --UGGCGCAGAAAAAAAAAGGACAAAAUGUCACAAAAGCGUGACGAUGUGUCGUAAAGUACUCAGACAAACACUAACCUCAA --.((...............((((...((((((......))))))...)))).....((......)).........)).... ( -12.60, z-score = -0.14, R) >droPer1.super_2858 1399 72 - 5027 ----------UGGAAAAAAGGACAAAAUGUCACAAAAGCGUGACGAUGUGUCGUAAAGUACUCAGACAAACACUAACCUCAA ----------.((.......((((...((((((......))))))...)))).....((......)).........)).... ( -13.30, z-score = -0.99, R) >droWil1.scaffold_180698 6693436 60 + 11422946 UCUUGAGUGGUAAAAAGG---ACAAAAUGUCACAAAAGCGUGACGAUGUGUCGUAAAGUACCC------------------- ........((((...(.(---(((...((((((......))))))...)))).)....)))).------------------- ( -16.40, z-score = -3.30, R) >droVir3.scaffold_13049 12563149 71 + 25233164 --UUGAGUGGC-AAAAGG---ACAAAAUGUCACAAAAGCGUGACGAUGUGUCGUAAAGUACCCAA-----CACUAACCCAAU --...((((..-.....(---(((...((((((......))))))...))))(((...)))....-----))))........ ( -15.10, z-score = -1.36, R) >droMoj3.scaffold_6680 9886357 72 - 24764193 --UUGAGUGGC-GAAAGG---ACAAAAUGUCACAAAAGCGUGACGAUGUGUCGUAAAGUACCCAAA----CACUAACCCAAA --...((((.(-....)(---(((...((((((......))))))...))))..............----))))........ ( -17.80, z-score = -2.18, R) >droGri2.scaffold_15110 1411713 70 - 24565398 --UUGAGUGG--AAAAGG---ACAAAAUGUCACAAAAGCGUGACGAUGUGUCGUAAAGUACCCAA-----CACUAGCCCAAA --...((((.--.....(---(((...((((((......))))))...))))(((...)))....-----))))........ ( -15.10, z-score = -1.46, R) >consensus __UUGGGCGGGGAAAAAA__GACAAAAUGUCACAAAAGCGUGACGAUGUGUCGUAAAGUACUCAGACAA_CACUAACCUCAA .....................(((...((((((......))))))...)))............................... ( -8.27 = -8.27 + 0.00)

| Location | 13,176,471 – 13,176,570 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 93.29 |
| Shannon entropy | 0.12102 |
| G+C content | 0.41350 |
| Mean single sequence MFE | -25.76 |
| Consensus MFE | -21.78 |
| Energy contribution | -23.18 |
| Covariance contribution | 1.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.85 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.745822 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
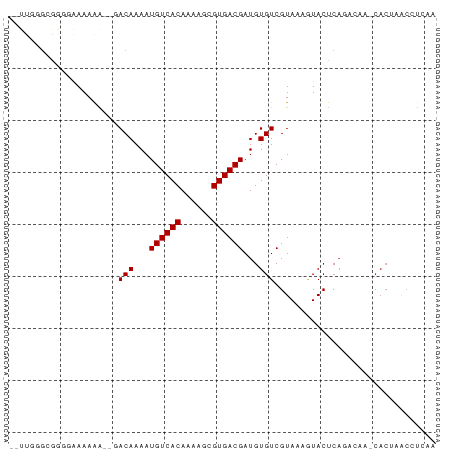
>dm3.chr3L 13176471 99 + 24543557 AAUGGGGUCUCGAAUGAUAUGGAAAAUGUCUUUUCAAAGACCUCGAAACUUGGCUUUUUGUACUUUUCACUGGUCCUUUAGCUUAUGCUUGGAUACUCU ...(((((((.(((.(((((.....)))))..)))..)))))))((((....((.....))...))))....((((...(((....))).))))..... ( -24.90, z-score = -2.42, R) >droSim1.chr3L 12571905 98 + 22553184 AAUGGGGUCUCGAAUGAUAUGGAAAAUGUCUUUUCAGAGACCUCGAAACUUGGCUUUU-GUACUUUUCACUGGUCCUUUAGCAUUUGCUUGGAUACCCU ...(((((((((((.(((((.....)))))..))).))))))))((((....((....-))...))))....((((...(((....))).))))..... ( -28.80, z-score = -2.68, R) >droSec1.super_0 5352229 98 + 21120651 AAUGGGGUCUCGAAUGAUAUGGAAAAUGUCUUUUCAGAGACCUCGAAACUUGGCUUUU-GUACUUUUCACUGUUCCUCUAGCUUCUGCUUGGAUACCCU ...(((((((((((.(((((.....)))))..))).))))))))((((....((....-))...)))).....(((...(((....))).)))...... ( -27.30, z-score = -2.51, R) >droYak2.chr3L 13263085 98 + 24197627 AAUGGGGUCUCGAAUGAUAUGGAAAAUGUCUUUCCAAAGACCUCGAACCUUGGCUUUU-GUACUUUUCACUGGUCCUGAAGCUUUUGCUUGGAUACCCU ..((((((((.(((.(((((.....))))).)))...))))))))..((..((((..(-(.......))..))))...((((....))))))....... ( -24.00, z-score = -1.07, R) >droEre2.scaffold_4784 13184968 98 + 25762168 AAUGGGGUCUCGAAUGAUAUGGAAAAUGUCUUUUCAGAGAGCUCGAACCUUGGCUAUU-GUACUUUUCACUGGUCCUAAAGCUGUUGCUUGGAUACCCU ..((((.(((((((.(((((.....)))))..))).)))).))))..((..(((((.(-(.......)).)))))...((((....))))))....... ( -23.80, z-score = -0.54, R) >consensus AAUGGGGUCUCGAAUGAUAUGGAAAAUGUCUUUUCAGAGACCUCGAAACUUGGCUUUU_GUACUUUUCACUGGUCCUUUAGCUUUUGCUUGGAUACCCU ...(((((((((((.(((((.....)))))..))).))))))))((((....((.....))...))))....((((...(((....))).))))..... (-21.78 = -23.18 + 1.40)



Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:22:56 2011