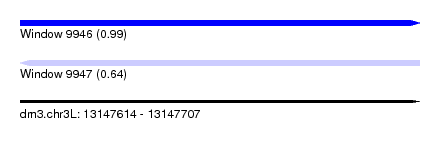
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 13,147,614 – 13,147,707 |
| Length | 93 |
| Max. P | 0.989807 |

| Location | 13,147,614 – 13,147,707 |
|---|---|
| Length | 93 |
| Sequences | 8 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 76.89 |
| Shannon entropy | 0.44689 |
| G+C content | 0.47497 |
| Mean single sequence MFE | -32.52 |
| Consensus MFE | -21.28 |
| Energy contribution | -22.75 |
| Covariance contribution | 1.47 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.39 |
| SVM RNA-class probability | 0.989807 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
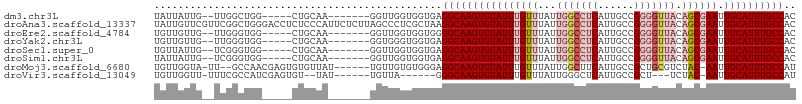
>dm3.chr3L 13147614 93 + 24543557 UAUUAUUG--UUGGCUGG-----CUGCAA-------GGUUGGUGGUGAGGCAAGUGUAUUUGUUUAUUGGCCUCAUUGCCGGGGUUACAGCGAAUUGCAUUUGCCAC ..((((..--(..(((..-----......-------)))..)..))))((((((((((((((((...(((((((......))))))).)))))).)))))))))).. ( -35.60, z-score = -2.97, R) >droAna3.scaffold_13337 10964150 107 - 23293914 UAUUGUUCGUUCGGCUGGGACCUCUCCCAUUCUCUUAGCCCUCGCUAAGGCAAGUGUAUUUGUUUAUUGGCCUCAUUGCCGGGGUUACAGCGAAUUGCAUUUGCCAC ............(..(((((....)))))..)..(((((....)))))((((((((((((((((...(((((((......))))))).)))))).)))))))))).. ( -36.60, z-score = -2.79, R) >droEre2.scaffold_4784 13156401 93 + 25762168 UGUUGUUG--UUGGGUGG-----CUGCAA-------GGUUGGUGGUGGGGCAAGUGUAUUUGUUUAUUGGCCUCAUUGCCGGGGUUACAGCGAAUUGCAUUUGCCAC ........--....((((-----((((((-------..((((..(((((((.((((........)))).)))))))..)))).((....))...)))))...))))) ( -31.80, z-score = -1.62, R) >droYak2.chr3L 13233074 93 + 24197627 UGUUGUUG--UUGGGUGG-----CUGCAA-------GGUGGGUGGUGAGGCAAGUGUAUUUGUUUAUUGGCCUCAUUGCCGGGGUUACAGCGAAUUGCAUUUGCCAC ........--....((((-----((((((-------..((((..(((((((.((((........)))).)))))))..))(......)..))..)))))...))))) ( -33.80, z-score = -2.29, R) >droSec1.super_0 5323559 93 + 21120651 UGUUAUUG--UCGGGUGG-----CUGCAA-------GGUUGGUGGUGAGGCAAGUGUAUUUGUUUAUUGGCCUCAUUGCCGGGGUUACAGCGAAUUGCAUUUGCCAC ........--....((((-----((((((-------..((((..(((((((.((((........)))).)))))))..)))).((....))...)))))...))))) ( -32.70, z-score = -2.03, R) >droSim1.chr3L 12541813 93 + 22553184 UAUUAUUG--UCGGGUGG-----CUGCAA-------GGUUGGUGGUGAGGCAAGUGUAUUUGUUUAUUGGCCUCAUUGCCGGGGUUACAGCGAAUUGCAUUUGCCAC ........--....((((-----((((((-------..((((..(((((((.((((........)))).)))))))..)))).((....))...)))))...))))) ( -32.70, z-score = -2.24, R) >droMoj3.scaffold_6680 9849600 97 - 24764193 UGUUGGUA-UU--GCCAACGAGUGUGUUAU------UGUUGUGUGGGAGGCAAGUGUAUUUGUUUAUUGGCUUCAUUGCCGCUGCGUCUAC-AAUUGCAUUUGCCAU ........-(.--.(((((((........)------))))).)..)..((((((((((.((((..((((((......))))....))..))-)).)))))))))).. ( -30.50, z-score = -1.61, R) >droVir3.scaffold_13049 12528601 88 + 25233164 UGUUGGUU-UUUCGCCAUCGAGUGU--UAU------UGUUA------GGGCAAGUGUAUUUGUUUAUUGGGCUCAUUGCCGCU---UCUAC-AAUUGCAUUUGCCAU .(.((((.-....)))).)......--...------.....------.((((((((((.((((.....((((.....))).).---...))-)).)))))))))).. ( -26.50, z-score = -2.56, R) >consensus UGUUGUUG__UUGGCUGG_____CUGCAA_______GGUUGGUGGUGAGGCAAGUGUAUUUGUUUAUUGGCCUCAUUGCCGGGGUUACAGCGAAUUGCAUUUGCCAC ................................................((((((((((((((((...(((((((......))))))).)))))).)))))))))).. (-21.28 = -22.75 + 1.47)

| Location | 13,147,614 – 13,147,707 |
|---|---|
| Length | 93 |
| Sequences | 8 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 76.89 |
| Shannon entropy | 0.44689 |
| G+C content | 0.47497 |
| Mean single sequence MFE | -21.65 |
| Consensus MFE | -10.70 |
| Energy contribution | -10.64 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.635978 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
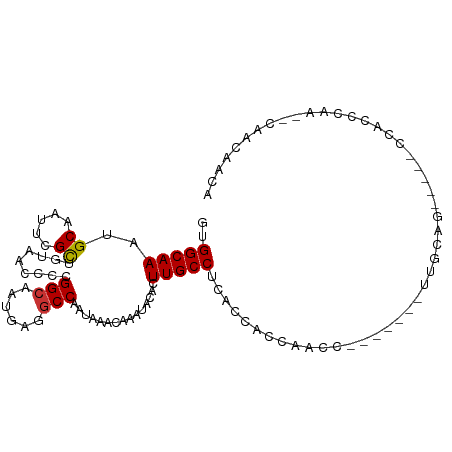
>dm3.chr3L 13147614 93 - 24543557 GUGGCAAAUGCAAUUCGCUGUAACCCCGGCAAUGAGGCCAAUAAACAAAUACACUUGCCUCACCACCAACC-------UUGCAG-----CCAGCCAA--CAAUAAUA .((((...(((((...((((......))))..((((((..................)))))).........-------))))).-----...)))).--........ ( -22.87, z-score = -2.09, R) >droAna3.scaffold_13337 10964150 107 + 23293914 GUGGCAAAUGCAAUUCGCUGUAACCCCGGCAAUGAGGCCAAUAAACAAAUACACUUGCCUUAGCGAGGGCUAAGAGAAUGGGAGAGGUCCCAGCCGAACGAACAAUA .((((.......((((.((....(((..((..((((((..................))))))))..)))...)).))))((((....)))).))))........... ( -26.57, z-score = -0.11, R) >droEre2.scaffold_4784 13156401 93 - 25762168 GUGGCAAAUGCAAUUCGCUGUAACCCCGGCAAUGAGGCCAAUAAACAAAUACACUUGCCCCACCACCAACC-------UUGCAG-----CCACCCAA--CAACAACA (((((...(((((...((((......))))..((.(((..................))).)).........-------))))))-----))))....--........ ( -19.17, z-score = -1.23, R) >droYak2.chr3L 13233074 93 - 24197627 GUGGCAAAUGCAAUUCGCUGUAACCCCGGCAAUGAGGCCAAUAAACAAAUACACUUGCCUCACCACCCACC-------UUGCAG-----CCACCCAA--CAACAACA (((((...(((((...((((......))))..((((((..................)))))).........-------))))))-----))))....--........ ( -23.97, z-score = -3.12, R) >droSec1.super_0 5323559 93 - 21120651 GUGGCAAAUGCAAUUCGCUGUAACCCCGGCAAUGAGGCCAAUAAACAAAUACACUUGCCUCACCACCAACC-------UUGCAG-----CCACCCGA--CAAUAACA (((((...(((((...((((......))))..((((((..................)))))).........-------))))))-----))))....--........ ( -23.97, z-score = -2.99, R) >droSim1.chr3L 12541813 93 - 22553184 GUGGCAAAUGCAAUUCGCUGUAACCCCGGCAAUGAGGCCAAUAAACAAAUACACUUGCCUCACCACCAACC-------UUGCAG-----CCACCCGA--CAAUAAUA (((((...(((((...((((......))))..((((((..................)))))).........-------))))))-----))))....--........ ( -23.97, z-score = -3.03, R) >droMoj3.scaffold_6680 9849600 97 + 24764193 AUGGCAAAUGCAAUU-GUAGACGCAGCGGCAAUGAAGCCAAUAAACAAAUACACUUGCCUCCCACACAACA------AUAACACACUCGUUGGC--AA-UACCAACA ..(((((.((...((-((....)))).(((......)))............)).)))))............------...........(((((.--..-..))))). ( -18.60, z-score = -0.88, R) >droVir3.scaffold_13049 12528601 88 - 25233164 AUGGCAAAUGCAAUU-GUAGA---AGCGGCAAUGAGCCCAAUAAACAAAUACACUUGCCC------UAACA------AUA--ACACUCGAUGGCGAAA-AACCAACA ..(((((.((...((-((...---...(((.....)))......))))...)).))))).------.....------...--......(.(((.....-..))).). ( -14.10, z-score = -0.32, R) >consensus GUGGCAAAUGCAAUUCGCUGUAACCCCGGCAAUGAGGCCAAUAAACAAAUACACUUGCCUCACCACCAACC_______UUGCAG_____CCACCCAA__CAACAACA ..(((((..((.....)).........(((......)))...............)))))................................................ (-10.70 = -10.64 + -0.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:22:52 2011