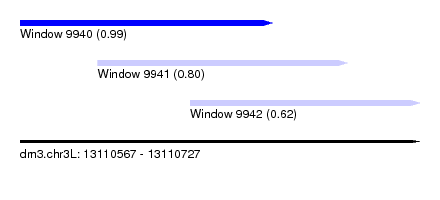
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 13,110,567 – 13,110,727 |
| Length | 160 |
| Max. P | 0.994902 |

| Location | 13,110,567 – 13,110,668 |
|---|---|
| Length | 101 |
| Sequences | 11 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 69.78 |
| Shannon entropy | 0.64090 |
| G+C content | 0.54006 |
| Mean single sequence MFE | -32.90 |
| Consensus MFE | -14.48 |
| Energy contribution | -14.08 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.40 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.75 |
| SVM RNA-class probability | 0.994902 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 13110567 101 + 24543557 ---------AGCAGCAGCAGAACAACAACACCCUGCGUCAGUUCACGCACUGAUCCUCGAC---CUAUCGACGCAGGCAGCUGAGCAUCUAUGCUUAGU-AGUCCGCUGCCCAA-- ---------....(((((.((..........(((((((((((......)))))...((((.---...))))))))))..((((((((....))))))))-..)).)))))....-- ( -35.70, z-score = -3.26, R) >droSim1.chr3L 12503920 101 + 22553184 ---------AGCAGCAGCAGAACAACAACACCCUGCGUCAGUUCACGCACUGAUCCUCGAC---CUAUCGACGCAGGCAGCUGAGCAUCUAUGCUUAGU-AGUCCGCUGCCCAA-- ---------....(((((.((..........(((((((((((......)))))...((((.---...))))))))))..((((((((....))))))))-..)).)))))....-- ( -35.70, z-score = -3.26, R) >droSec1.super_0 5286307 101 + 21120651 ---------AGCAGCAGCAGAACAACAACACCCUGCGUCAGUUCACGCACUGAUCCUCGAC---CUAUCGACGCAGGCAGCUGAGCAUCUAUGCUUAGU-AGUCCGCUGCCCAA-- ---------....(((((.((..........(((((((((((......)))))...((((.---...))))))))))..((((((((....))))))))-..)).)))))....-- ( -35.70, z-score = -3.26, R) >droEre2.scaffold_4784 13118733 104 + 25762168 ------AGCAGCAGCAGCAGAACAACAACACACUGCGUCAGUUCACGCACUGAUCCUCGAC---CUAUAGACGCAGGCAGCUGAGCAUCUAUGCUUAGU-AGUCCGCUGCCCAA-- ------.(((((.((.((((............))))((((((......)))))).......---........)).(((.((((((((....))))))))-.))).)))))....-- ( -33.90, z-score = -1.95, R) >droAna3.scaffold_13337 10929879 104 - 23293914 ------ACCAGCAGCAGCAGAACAACAACACCCUGCGCCAGUUCACGCACUGAUCCUCGAC---CUACCGACGCAGGCAGCUGAGUAUCUUUGCUUAGC-CCCCUGCUGCCCCC-- ------.......(((((((...........((((((.((((......))))....(((..---....)))))))))..((((((((....))))))))-...)))))))....-- ( -36.70, z-score = -4.66, R) >dp4.chrXR_group6 5374892 94 + 13314419 --------------------AACAACAACACCCUGCGGCAGUUCACCCACUGAUCCACGGUUGACUAUCGACGCAGGCAGCUGAGCAUCUACGCUUAGUUCGGCCGCUGUCCAA-- --------------------.........(((.((.((((((......)))).)))).)))........(((((.(((.(((((((......)))))))...))))).)))...-- ( -26.40, z-score = -0.74, R) >droPer1.super_257 23362 94 + 32565 --------------------AACAACAACACCCUGCGGCAGUUCACCCACUGAUCCACGGUUGACUAUCGACGCAGGCAGCUGAGCAUCUACGCUUAGUUCGGCCGCUGUCCAA-- --------------------.........(((.((.((((((......)))).)))).)))........(((((.(((.(((((((......)))))))...))))).)))...-- ( -26.40, z-score = -0.74, R) >droWil1.scaffold_180698 6603231 104 + 11422946 CGCUGCAGUAGCUACAUCAA-AUAAUAAUACAAUGCGACAAUUUAGCCACUGAUCUUCAAC---CUAUCGACGACGGAAUCUAAGCAUAUAUGCUUAGAUUCGCUUAA-------- .((..((((.((((......-...((......)).........)))).)))).........---............(((((((((((....)))))))))))))....-------- ( -23.07, z-score = -2.24, R) >droVir3.scaffold_13049 12485598 111 + 25233164 --GCCAGCACACAACAAUAAUACG--AGCACUAUGCGCCAGUUUACGCACUGAUCCUCGGAGCAUUAUCGUCGCAGACAGCUGAGCAUUUAUGCUUAGUCGU-UCGUGUCGUUGUC --...........((((..(((((--(((....((((.(.((....((.(((.....))).))...)).).))))....((((((((....)))))))).))-))))))..)))). ( -32.90, z-score = -1.47, R) >droMoj3.scaffold_6680 9798430 111 - 24764193 ---AACAACAACAGCAGCAAUGCG--AGCACCAUGCGCCAGUUCACGCACUGAUCCUCGGAGCACUAUCGUCGCAGGCAGCUGAGCAUCUAUGCUUAGUCGUGUCGUGUCGUUGUC ---....(((((.(((....)))(--(.(((..((((.(.((....((.(((.....))).))...)).).))))((((((((((((....))))))))..)))))))))))))). ( -34.80, z-score = -0.12, R) >droGri2.scaffold_15110 23094477 99 + 24565398 ------------GCCAGCGA--CG--AACACCUUGCGGCAAUUCACGCACUGAUCCUCGGAGCACUAUCGUCGCAGGCAGCUGAGCAUUUUUGCUUAGUUCC-UCGUCGCGUUGCC ------------((..((((--((--(......((((((.((....((.(((.....))).))...)).))))))((.(((((((((....)))))))))))-)))))))...)). ( -40.60, z-score = -3.58, R) >consensus _________AGCAGCAGCAGAACAACAACACCCUGCGUCAGUUCACGCACUGAUCCUCGAC___CUAUCGACGCAGGCAGCUGAGCAUCUAUGCUUAGU_AGGCCGCUGCCCAA__ .................................((((.((((......)))).....(((.......))).))))((..(((((((......)))))))....))........... (-14.48 = -14.08 + -0.40)
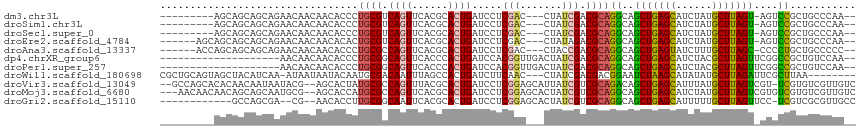
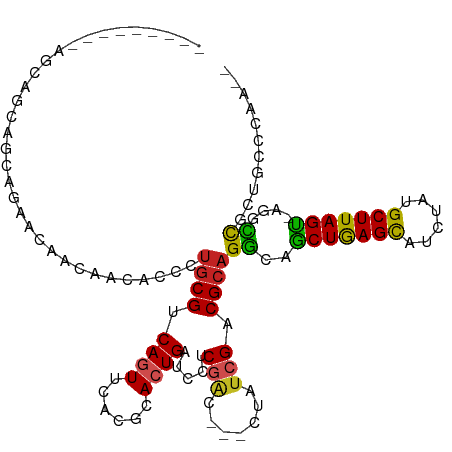
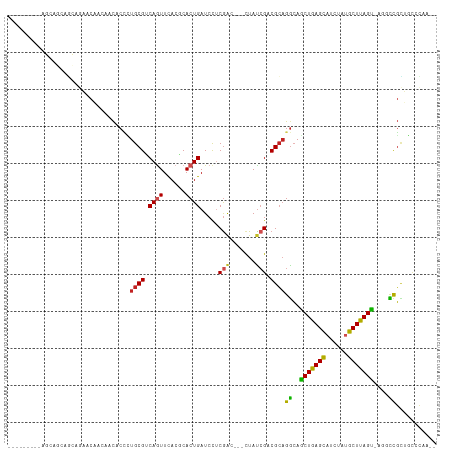
| Location | 13,110,598 – 13,110,698 |
|---|---|
| Length | 100 |
| Sequences | 12 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 73.74 |
| Shannon entropy | 0.54394 |
| G+C content | 0.53726 |
| Mean single sequence MFE | -24.79 |
| Consensus MFE | -13.46 |
| Energy contribution | -13.41 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.801895 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 13110598 100 + 24543557 GUUCACGCACUGAUCCUCGAC---CUAUCGACGCAGGCAGCUGAGCAUCUAUGCUUAGUA---GUCCGCUGCCCAACACUGUGAUGCCAAA-ACCCCCAAGAAGCCC------- .....((((.((....((((.---...)))).((.(((.((((((((....)))))))).---))).)).......)).))))........-...............------- ( -23.40, z-score = -0.94, R) >droSim1.chr3L 12503951 100 + 22553184 GUUCACGCACUGAUCCUCGAC---CUAUCGACGCAGGCAGCUGAGCAUCUAUGCUUAGUA---GUCCGCUGCCCAACACUGUGAUGCCAAA-ACCCCCAAAAAGCCC------- .....((((.((....((((.---...)))).((.(((.((((((((....)))))))).---))).)).......)).))))........-...............------- ( -23.40, z-score = -1.46, R) >droSec1.super_0 5286338 100 + 21120651 GUUCACGCACUGAUCCUCGAC---CUAUCGACGCAGGCAGCUGAGCAUCUAUGCUUAGUA---GUCCGCUGCCCAACACUGUGAUGCCAAA-ACCCCCAAAAAGCCC------- .....((((.((....((((.---...)))).((.(((.((((((((....)))))))).---))).)).......)).))))........-...............------- ( -23.40, z-score = -1.46, R) >droYak2.chr3L 13194464 100 + 24197627 GUUCACGCACUGAUCCUCGAC---CUAUCGACGCAGGCAGCUGAGCAUCUUUGCUUAGUA---GUCCGCUGCCCAACACUGUGAUGCCAAA-ACCCCCAAAAAGCCC------- .....((((.((....((((.---...)))).((.(((.((((((((....)))))))).---))).)).......)).))))........-...............------- ( -24.20, z-score = -1.61, R) >droEre2.scaffold_4784 13118767 100 + 25762168 GUUCACGCACUGAUCCUCGAC---CUAUAGACGCAGGCAGCUGAGCAUCUAUGCUUAGUA---GUCCGCUGCCCAACACUGUGAUGCCAAA-ACCCCCAAAAAACAC------- .....((((.((....((...---.....)).((.(((.((((((((....)))))))).---))).)).......)).))))........-...............------- ( -20.70, z-score = -0.86, R) >droAna3.scaffold_13337 10929913 107 - 23293914 GUUCACGCACUGAUCCUCGAC---CUACCGACGCAGGCAGCUGAGUAUCUUUGCUUAGCC---CCCUGCUGCCCCCCACUGUGAUGCCUGG-UCCUGCCCCACCCUCACAUCCU ......(((.......(((..---....))).(((((..((((((((....)))))))).---.)))))))).......(((((.(..(((-.......)))..)))))).... ( -29.60, z-score = -2.83, R) >dp4.chrXR_group6 5374912 106 + 13314419 GUUCACCCACUGAUCCACGGUUGACUAUCGACGCAGGCAGCUGAGCAUCUACGCUUAGUUC--GGCCGCUGUCCAACACUGUGAUGCCAAA-ACCAACCCCCAGCCCCA----- .........(((......((((...(((((((((.(((.(((((((......)))))))..--.))))).)))((....)).))))....)-)))......))).....----- ( -24.50, z-score = -0.08, R) >droPer1.super_257 23382 106 + 32565 GUUCACCCACUGAUCCACGGUUGACUAUCGACGCAGGCAGCUGAGCAUCUACGCUUAGUUC--GGCCGCUGUCCAACACUGUGAUGCCAAA-ACCAACCCCCAGCCCCA----- .........(((......((((...(((((((((.(((.(((((((......)))))))..--.))))).)))((....)).))))....)-)))......))).....----- ( -24.50, z-score = -0.08, R) >droWil1.scaffold_180698 6603270 98 + 11422946 AUUUAGCCACUGAUCUUCAAC---CUAUCGACGACGGAAUCUAAGCAUAUAUGCUUAGAU--------UCGCUUAACACUGUGAUGCCAAACAAUUUCCCCGCACUCCC----- .....(((((.(.((.((...---.....)).))..(((((((((((....)))))))))--------))........).)))..))......................----- ( -17.50, z-score = -2.09, R) >droVir3.scaffold_13049 12485634 105 + 25233164 GUUUACGCACUGAUCCUCGGAGCAUUAUCGUCGCAGACAGCUGAGCAUUUAUGCUUAGUCGU-UCGUGUCGUUGUCAACUGUGAUGCCAAA-AUUUGUAAAACCUCC------- .(((((...(((.....))).(((((((((.(((..((.((((((((....)))))))).))-..))).)).........)))))))....-....)))))......------- ( -28.20, z-score = -1.62, R) >droMoj3.scaffold_6680 9798465 103 - 24764193 GUUCACGCACUGAUCCUCGGAGCACUAUCGUCGCAGGCAGCUGAGCAUCUAUGCUUAGUCGUGUCGUGUCGUUGUCAACUGUGAUGCCAAA-ACUCCACCCGCC---------- ......((...(.....)((((......((((((((((.((((((((....)))))))).))(.((......)).)..)))))))).....-.))))....)).---------- ( -28.40, z-score = -0.16, R) >droGri2.scaffold_15110 23094501 98 + 24565398 AUUCACGCACUGAUCCUCGGAGCACUAUCGUCGCAGGCAGCUGAGCAUUUUUGCUUAGUUCC-UCGUCGCGUUGCCAACUGUGAUGCCAAA-UUCUGUAA-------------- ......((.(((.....))).))......(.((.(((.(((((((((....)))))))))))-))).)((((..(.....)..))))....-........-------------- ( -29.70, z-score = -1.57, R) >consensus GUUCACGCACUGAUCCUCGAC___CUAUCGACGCAGGCAGCUGAGCAUCUAUGCUUAGUA___GUCCGCUGCCCAACACUGUGAUGCCAAA_ACCCCCAAAAAGCCC_______ ..((((...(((.....(((.......)))...)))...((((((((....)))))))).....................)))).............................. (-13.46 = -13.41 + -0.05)
| Location | 13,110,635 – 13,110,727 |
|---|---|
| Length | 92 |
| Sequences | 7 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 74.57 |
| Shannon entropy | 0.48677 |
| G+C content | 0.53139 |
| Mean single sequence MFE | -12.40 |
| Consensus MFE | -7.41 |
| Energy contribution | -7.41 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.06 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.621176 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
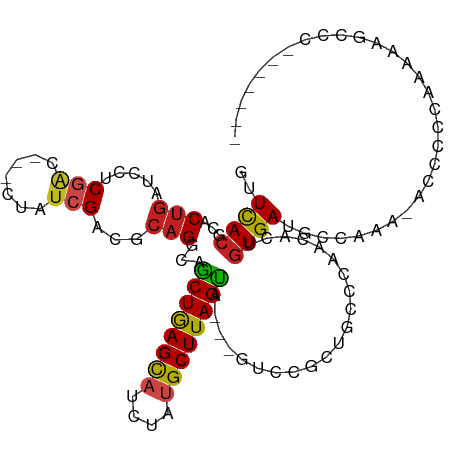
>dm3.chr3L 13110635 92 + 24543557 CUGAGCAUCUAUGCUUAGUAGU---CCGCUGCCCAACACUGUGAUGCCAAAACCC------CCAAGAAGCCCACUAAAACCACUCACUCCCCUC-CAGACAC (((((((....)))))))((((---..(((...........((....))......------......)))..))))..................-....... ( -11.60, z-score = -0.48, R) >droSim1.chr3L 12503988 92 + 22553184 CUGAGCAUCUAUGCUUAGUAGU---CCGCUGCCCAACACUGUGAUGCCAAAACCC------CCAAAAAGCCCACUAAAACCACUCCCUCCCCUC-CAGAGAC (((((((....)))))))((((---..(((...........((....))......------......)))..))))..........(((.....-..))).. ( -12.60, z-score = -1.36, R) >droSec1.super_0 5286375 92 + 21120651 CUGAGCAUCUAUGCUUAGUAGU---CCGCUGCCCAACACUGUGAUGCCAAAACCC------CCAAAAAGCCCACUAAAACCACUCCCUCCCCUC-CAGAGAC (((((((....)))))))((((---..(((...........((....))......------......)))..))))..........(((.....-..))).. ( -12.60, z-score = -1.36, R) >droYak2.chr3L 13194501 92 + 24197627 CUGAGCAUCUUUGCUUAGUAGU---CCGCUGCCCAACACUGUGAUGCCAAAACCC------CCAAAAAGCCCACUAAAACCACUCCCUCCCCUC-CAGAGAC (((((((....)))))))((((---..(((...........((....))......------......)))..))))..........(((.....-..))).. ( -13.40, z-score = -1.42, R) >droEre2.scaffold_4784 13118804 92 + 25762168 CUGAGCAUCUAUGCUUAGUAGU---CCGCUGCCCAACACUGUGAUGCCAAAACCC------CCAAAAAACACACUAAAACCACUCCCUCCCCUC-CAGAGAC .((.(((((..((....((((.---...))))....))....)))))))......------.........................(((.....-..))).. ( -11.00, z-score = -1.07, R) >droVir3.scaffold_13049 12485674 90 + 25233164 CUGAGCAUUUAUGCUUAGUCGU-UCGUGUCGUUGUCAACUGUGAUGCCAAAAUUUGUAAAACCUCCCUGUCCCCC------GUCCCCCUCCUCCACA----- (((((((....)))))))....-..((((((..(....)..))))))............................------................----- ( -11.50, z-score = -1.31, R) >droMoj3.scaffold_6680 9798505 102 - 24764193 CUGAGCAUCUAUGCUUAGUCGUGUCGUGUCGUUGUCAACUGUGAUGCCAAAACUCCACCCGCCCUACCGCCCUCCCCGAUUGCUCCCCUCUCCCACAACCAC (((((((....)))))))..(((..((((((..(....)..)))))).............((......)).......................)))...... ( -14.10, z-score = -0.37, R) >consensus CUGAGCAUCUAUGCUUAGUAGU___CCGCUGCCCAACACUGUGAUGCCAAAACCC______CCAAAAAGCCCACUAAAACCACUCCCUCCCCUC_CAGAGAC (((((((....))))))).................................................................................... ( -7.41 = -7.41 + 0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:22:48 2011