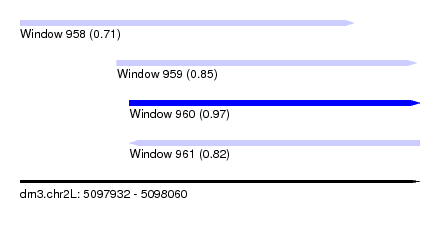
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 5,097,932 – 5,098,060 |
| Length | 128 |
| Max. P | 0.968662 |

| Location | 5,097,932 – 5,098,039 |
|---|---|
| Length | 107 |
| Sequences | 10 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 75.43 |
| Shannon entropy | 0.53875 |
| G+C content | 0.55516 |
| Mean single sequence MFE | -36.26 |
| Consensus MFE | -21.11 |
| Energy contribution | -20.66 |
| Covariance contribution | -0.45 |
| Combinations/Pair | 1.59 |
| Mean z-score | -0.97 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.708670 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
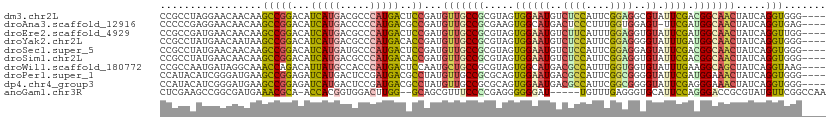
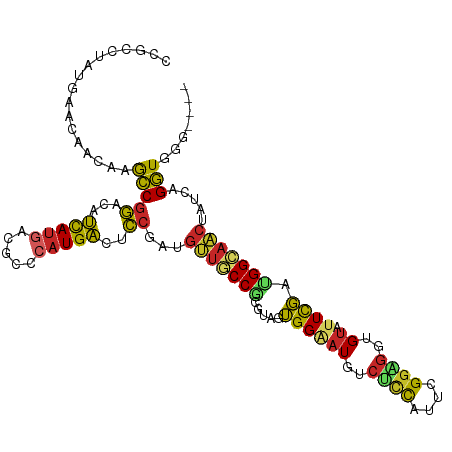
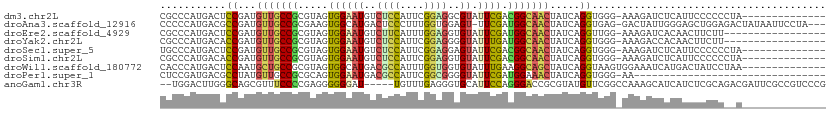
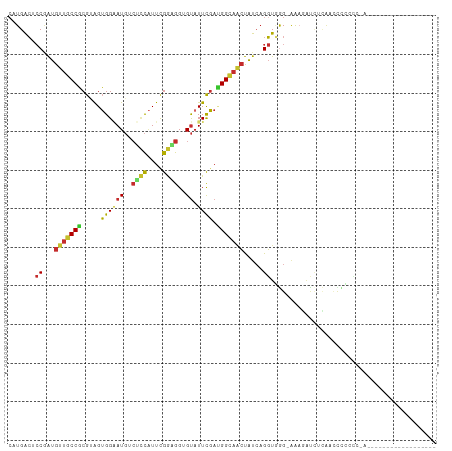
>dm3.chr2L 5097932 107 + 23011544 CCGCCUAGGAACAACAAGCCGGACAUCAUGACGCCCAUGACUCCGAUGUUGCCGCGUAGUGGAAUGUCUCCAUUCGGAGGCGUAUUCGACGGCAACUAUCAGGUGGG---- ((((((..............(((..(((((.....))))).)))((((((((((((.(((...((((((((....))))))))))))).))))))).))))))))).---- ( -44.40, z-score = -3.16, R) >droAna3.scaffold_12916 6112083 106 - 16180835 CCCCCGAGGAACAACAAGCCGGACAUCAUGACCCCCAUGACGCCGAUGUUGCCGCGAAGUGGCAUGACUCCCUUUGGUGGAGU-UUCGAUGGCAACUAUCAGGUGAG---- ...(((.............)))...(((((.....)))))(((((((((((((((((........(((((((....).)))))-)))).))))))).))).))))..---- ( -33.02, z-score = -0.82, R) >droEre2.scaffold_4929 5178267 107 + 26641161 CCGCCGAUGAACAACAAGCCGGACAUCAUGACGCCCAUGACUCCGAUGUUGCCGCGUAGUGGAAUGUCUUCAUUUGGAGGUGUAUUCGAUGGCAACUAUCAGGUUGG---- (((((((((..........((((..(((((.....))))).))))..(((((((((.(((...((..((((....))))..))))))).))))))))))).)).)))---- ( -31.70, z-score = -0.14, R) >droYak2.chr2L 11898204 107 - 22324452 CCGCCUAUGAACAAUAAGCCGGACAUCAUGACGCCCAUGACACCGAUGUUGCCGCGUAGUGGAAUGUCUCCAUUCGGAGGGGUAUUUGAUGGCAACUAUCAGGUGGG---- ((((((..............((...(((((.....)))))..))((((((((((((.(((...((.(((((....))))).))))))).))))))).))))))))).---- ( -35.20, z-score = -0.81, R) >droSec1.super_5 3184606 107 + 5866729 CCGCCUAUGAACAACAAGCCGGACAUCAUGAUGCCCAUGACUCCGAUGUUGCCGCGUAGUGGAAUGUCUCCAUUCGGAGGAGUAUUCGACGGCAACUAUCAGGUGGG---- ((((((..............(((..(((((.....))))).)))((((((((((((.((((.....(((((....)))))..)))))).))))))).))))))))).---- ( -40.30, z-score = -2.47, R) >droSim1.chr2L 4911543 107 + 22036055 CCGCCUAUGAACAACAAGCCGGACAUCAUGACGCCCAUGACACCGAUGUUGCCGCGUAGUGGAAUGUCUCCAUUCGGAGGUGUAUUCGACGGCAACUAUCAGGUGGG---- ((((((..............((...(((((.....)))))..))((((((((((((.(((...((..((((....))))..))))))).))))))).))))))))).---- ( -39.50, z-score = -2.05, R) >droWil1.scaffold_180772 1202845 107 + 8906247 CCGCCAAUGAUAGGCAAACCAGACAUUAUGCCACCCAUGACUCCAAUGCUGCCGCGUAGUGGCAUGACGCCAUUUGGUGGUGUAUUUGAAGGCAGCUAUCAGGUAAG---- ..(((..((((((((....((((...(((((((((..........((((....))))((((((.....)))))).)))))))))))))...))..)))))))))...---- ( -31.80, z-score = -0.32, R) >droPer1.super_1 5978488 107 - 10282868 CCAUACAUCGGGAUGAAGCCGGAGAUCAUGACUCCGAUGACGCCUAUGUUGCCGCGCAGUGGAAUGACGCCAUUCGGCGGGGUAUUCGAUGGAAACUAUCAGGUGGG---- ((...((((.((......))((((.......)))))))).(((((....((((.(((....(((((....))))).))).))))...((((....).))))))))))---- ( -36.30, z-score = -0.58, R) >dp4.chr4_group3 8873209 107 - 11692001 CCAUACAUCGGGAUGAAGCCGGAGAUCAUGACUCCGAUGACGCCUAUGUUGCCGCGCAGUGGAAUGACGCCAUUCGGCGGGGUAUUCGAGGGAAACUAUCAGGUGGG---- ((...((((.((......))((((.......)))))))).(((((....((((.(((....(((((....))))).))).))))...((((....)).)))))))))---- ( -35.80, z-score = -0.36, R) >anoGam1.chr3R 25762098 103 + 53272125 CUCGAAGCCGGCGAUGAAACGCA-ACCACGGUGGACUUGG--GCAGCGUUUCCCCGAGGGGGGAU-----UGUUUGAGGGUGCAUUCCAGGGACCGCGUAUGUUCGGCCAA ......(((((((......))).-......((((.(((((--(((.(.(((((((....))))).-----.....)).).)))...)))))..)))).......))))... ( -34.60, z-score = 1.03, R) >consensus CCGCCUAUGAACAACAAGCCGGACAUCAUGACGCCCAUGACUCCGAUGUUGCCGCGUAGUGGAAUGUCUCCAUUCGGAGGUGUAUUCGAUGGCAACUAUCAGGUGGG____ .................(((((...(((((.....)))))..))...(((((((.....((((((..((((....))))..)).)))).))))))).....)))....... (-21.11 = -20.66 + -0.45)
| Location | 5,097,963 – 5,098,059 |
|---|---|
| Length | 96 |
| Sequences | 9 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 65.22 |
| Shannon entropy | 0.71890 |
| G+C content | 0.53300 |
| Mean single sequence MFE | -31.01 |
| Consensus MFE | -14.52 |
| Energy contribution | -14.11 |
| Covariance contribution | -0.41 |
| Combinations/Pair | 1.63 |
| Mean z-score | -0.86 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.847571 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 5097963 96 + 23011544 CGCCCAUGACUCCGAUGUUGCCGCGUAGUGGAAUGUCUCCAUUCGGAGGCGUAUUCGACGGCAACUAUCAGGUGGG-AAAGAUCUCAUUCCCCCCUA-------------- .............((((((((((((.(((...((((((((....))))))))))))).))))))).)))(((.(((-((.(....).))))).))).-------------- ( -37.70, z-score = -2.86, R) >droAna3.scaffold_12916 6112114 106 - 16180835 CCCCCAUGACGCCGAUGUUGCCGCGAAGUGGCAUGACUCCCUUUGGUGGAGU-UUCGAUGGCAACUAUCAGGUGAG-GACUAUUGGGAGCUGGAGACUAUAAUUCCUA--- .........(((((((((((((((((........(((((((....).)))))-)))).))))))).))).))))..-......((((((..(....).....))))))--- ( -32.50, z-score = -0.49, R) >droEre2.scaffold_4929 5178298 93 + 26641161 CGCCCAUGACUCCGAUGUUGCCGCGUAGUGGAAUGUCUUCAUUUGGAGGUGUAUUCGAUGGCAACUAUCAGGUUGG-AAAGAUCACAACUUCUU----------------- .............((((((((((((.(((...((..((((....))))..))))))).))))))).)))(((((((-(....)).))))))...----------------- ( -23.70, z-score = 0.01, R) >droYak2.chr2L 11898235 93 - 22324452 CGCCCAUGACACCGAUGUUGCCGCGUAGUGGAAUGUCUCCAUUCGGAGGGGUAUUUGAUGGCAACUAUCAGGUGGG-AAAGACCACAACUUCUU----------------- .((((........((((((.((((...))))))))))(((....))).))))..((((((....).)))))((((.-.....))))........----------------- ( -26.70, z-score = -0.02, R) >droSec1.super_5 3184637 96 + 5866729 UGCCCAUGACUCCGAUGUUGCCGCGUAGUGGAAUGUCUCCAUUCGGAGGAGUAUUCGACGGCAACUAUCAGGUGGG-AAAGAUCUCAUUCCCCCCUA-------------- .............((((((((((((.((((.....(((((....)))))..)))))).))))))).)))(((.(((-((.(....).))))).))).-------------- ( -33.60, z-score = -1.77, R) >droSim1.chr2L 4911574 96 + 22036055 CGCCCAUGACACCGAUGUUGCCGCGUAGUGGAAUGUCUCCAUUCGGAGGUGUAUUCGACGGCAACUAUCAGGUGGG-AAAGAUCUCAUUCCCCCCUA-------------- .............((((((((((((.(((...((..((((....))))..))))))).))))))).)))(((.(((-((.(....).))))).))).-------------- ( -34.50, z-score = -1.97, R) >droWil1.scaffold_180772 1202876 97 + 8906247 CACCCAUGACUCCAAUGCUGCCGCGUAGUGGCAUGACGCCAUUUGGUGGUGUAUUUGAAGGCAGCUAUCAGGUAAGUGGAAAUCAUGACUAUCCUAA-------------- ....(((((.((((..((((((((......))...(((((((...))))))).......))))))...........))))..)))))..........-------------- ( -28.92, z-score = -0.69, R) >droPer1.super_1 5978519 78 - 10282868 CUCCGAUGACGCCUAUGUUGCCGCGCAGUGGAAUGACGCCAUUCGGCGGGGUAUUCGAUGGAAACUAUCAGGUGGG-AA-------------------------------- .(((.....(((((....((((.(((....(((((....))))).))).))))...((((....).))))))))))-).-------------------------------- ( -25.70, z-score = -0.52, R) >anoGam1.chr3R 25762128 104 + 53272125 --UGGACUUGGGCAGCGUUUCCCCGAGGGGGGAU-----UGUUUGAGGGUGCAUUCCAGGGACCGCGUAUGUUCGGCCAAAGCAUCAUCUCGCAGACGAUUCGCCGUCCCG --.((((...(((.......(((....)))((((-----((((((..(((((.......((.(((........)))))...)))))......))))))))))))))))).. ( -35.80, z-score = 0.56, R) >consensus CGCCCAUGACUCCGAUGUUGCCGCGUAGUGGAAUGUCUCCAUUCGGAGGUGUAUUCGAUGGCAACUAUCAGGUGGG_AAAGAUCUCAACCCCCC__A______________ ...........((...(((((((.....((((((..((((....))))..)).)))).))))))).....))....................................... (-14.52 = -14.11 + -0.41)

| Location | 5,097,967 – 5,098,060 |
|---|---|
| Length | 93 |
| Sequences | 9 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 64.76 |
| Shannon entropy | 0.71403 |
| G+C content | 0.51749 |
| Mean single sequence MFE | -30.97 |
| Consensus MFE | -13.91 |
| Energy contribution | -13.88 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.60 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.80 |
| SVM RNA-class probability | 0.968662 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
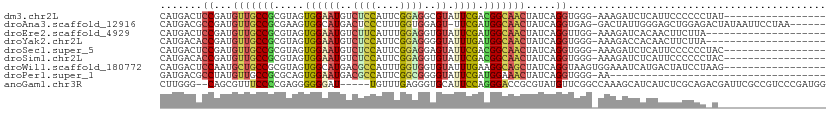
>dm3.chr2L 5097967 93 + 23011544 CAUGACUCCGAUGUUGCCGCGUAGUGGAAUGUCUCCAUUCGGAGGCGUAUUCGACGGCAACUAUCAGGUGGG-AAAGAUCUCAUUCCCCCCUAU----------------- .........((((((((((((.(((...((((((((....))))))))))))).))))))).)))(((.(((-((.(....).))))).)))..----------------- ( -37.70, z-score = -3.45, R) >droAna3.scaffold_12916 6112118 103 - 16180835 CAUGACGCCGAUGUUGCCGCGAAGUGGCAUGACUCCCUUUGGUGGAGU-UUCGAUGGCAACUAUCAGGUGAG-GACUAUUGGGAGCUGGAGACUAUAAUUCCUAA------ .....(((((((((((((((((........(((((((....).)))))-)))).))))))).))).))))..-.....(((((((..(....).....)))))))------ ( -33.50, z-score = -1.55, R) >droEre2.scaffold_4929 5178302 90 + 26641161 CAUGACUCCGAUGUUGCCGCGUAGUGGAAUGUCUUCAUUUGGAGGUGUAUUCGAUGGCAACUAUCAGGUUGG-AAAGAUCACAACUUCUUA-------------------- .........((((((((((((.(((...((..((((....))))..))))))).))))))).)))(((((((-(....)).))))))....-------------------- ( -23.70, z-score = -0.64, R) >droYak2.chr2L 11898239 90 - 22324452 CAUGACACCGAUGUUGCCGCGUAGUGGAAUGUCUCCAUUCGGAGGGGUAUUUGAUGGCAACUAUCAGGUGGG-AAAGACCACAACUUCUUA-------------------- .....((((((((((((((((.(((...((.(((((....))))).))))))).))))))).))).))))((-.....))...........-------------------- ( -26.00, z-score = -0.61, R) >droSec1.super_5 3184641 93 + 5866729 CAUGACUCCGAUGUUGCCGCGUAGUGGAAUGUCUCCAUUCGGAGGAGUAUUCGACGGCAACUAUCAGGUGGG-AAAGAUCUCAUUCCCCCCUAC----------------- .........((((((((((((.((((.....(((((....)))))..)))))).))))))).)))(((.(((-((.(....).))))).)))..----------------- ( -33.60, z-score = -2.21, R) >droSim1.chr2L 4911578 93 + 22036055 CAUGACACCGAUGUUGCCGCGUAGUGGAAUGUCUCCAUUCGGAGGUGUAUUCGACGGCAACUAUCAGGUGGG-AAAGAUCUCAUUCCCCCCUAC----------------- .........((((((((((((.(((...((..((((....))))..))))))).))))))).)))(((.(((-((.(....).))))).)))..----------------- ( -34.50, z-score = -2.51, R) >droWil1.scaffold_180772 1202880 94 + 8906247 CAUGACUCCAAUGCUGCCGCGUAGUGGCAUGACGCCAUUUGGUGGUGUAUUUGAAGGCAGCUAUCAGGUAAGUGGAAAUCAUGACUAUCCUAAG----------------- (((((.((((..((((((((......))...(((((((...))))))).......))))))...........))))..)))))...........----------------- ( -28.62, z-score = -1.15, R) >droPer1.super_1 5978523 74 - 10282868 GAUGACGCCUAUGUUGCCGCGCAGUGGAAUGACGCCAUUCGGCGGGGUAUUCGAUGGAAACUAUCAGGUGGG-AA------------------------------------ .....(((((....((((.(((....(((((....))))).))).))))...((((....).))))))))..-..------------------------------------ ( -25.40, z-score = -1.26, R) >anoGam1.chr3R 25762132 104 + 53272125 CUUGGG--CAGCGUUUCCCCGAGGGGGGAU-----UGUUUGAGGGUGCAUUCCAGGGACCGCGUAUGUUCGGCCAAAGCAUCAUCUCGCAGACGAUUCGCCGUCCCGAUGG .(((((--(.((....(((....)))((((-----((((((..(((((.......((.(((........)))))...)))))......)))))))))))).).)))))... ( -35.70, z-score = 0.54, R) >consensus CAUGACUCCGAUGUUGCCGCGUAGUGGAAUGUCUCCAUUCGGAGGUGUAUUCGAUGGCAACUAUCAGGUGGG_AAAGAUCUCAACCCCCCC_A__________________ .....(.((...(((((((.....((((((..((((....))))..)).)))).))))))).....)).)......................................... (-13.91 = -13.88 + -0.03)
| Location | 5,097,967 – 5,098,060 |
|---|---|
| Length | 93 |
| Sequences | 9 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 64.76 |
| Shannon entropy | 0.71403 |
| G+C content | 0.51749 |
| Mean single sequence MFE | -26.18 |
| Consensus MFE | -10.23 |
| Energy contribution | -11.27 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.818363 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
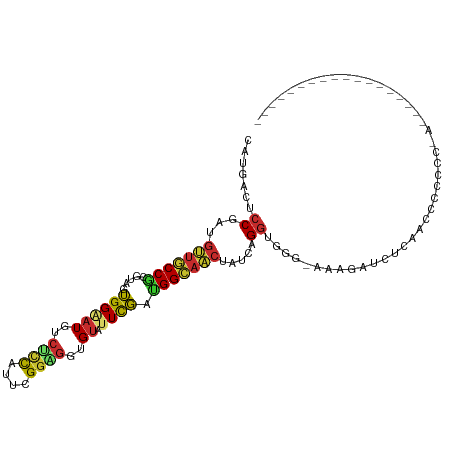
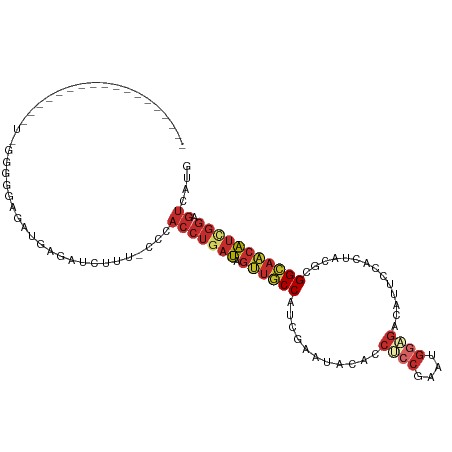
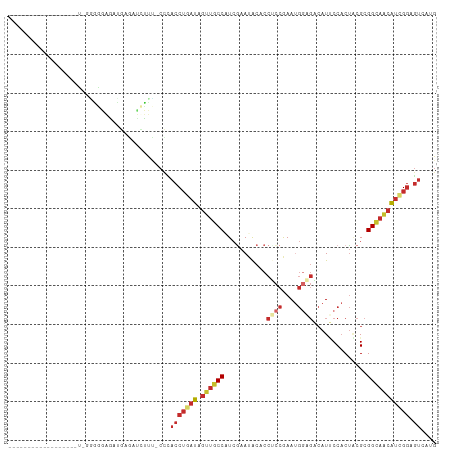
>dm3.chr2L 5097967 93 - 23011544 -----------------AUAGGGGGGAAUGAGAUCUUU-CCCACCUGAUAGUUGCCGUCGAAUACGCCUCCGAAUGGAGACAUUCCACUACGCGGCAACAUCGGAGUCAUG -----------------...((((((((........))-))).)))(((.((((((((((....)).....(((((....)))))......)))))))))))......... ( -32.20, z-score = -2.03, R) >droAna3.scaffold_12916 6112118 103 + 16180835 ------UUAGGAAUUAUAGUCUCCAGCUCCCAAUAGUC-CUCACCUGAUAGUUGCCAUCGAA-ACUCCACCAAAGGGAGUCAUGCCACUUCGCGGCAACAUCGGCGUCAUG ------..((((.(((((((.....)))....))))))-)).(((((((.((((((......-(((((.......)))))...((......))))))))))))).)).... ( -25.00, z-score = -1.28, R) >droEre2.scaffold_4929 5178302 90 - 26641161 --------------------UAAGAAGUUGUGAUCUUU-CCAACCUGAUAGUUGCCAUCGAAUACACCUCCAAAUGAAGACAUUCCACUACGCGGCAACAUCGGAGUCAUG --------------------...((.((((.((....)-)))))(((((.((((((...((.......)).....(((....)))........)))))))))))..))... ( -17.50, z-score = -0.72, R) >droYak2.chr2L 11898239 90 + 22324452 --------------------UAAGAAGUUGUGGUCUUU-CCCACCUGAUAGUUGCCAUCAAAUACCCCUCCGAAUGGAGACAUUCCACUACGCGGCAACAUCGGUGUCAUG --------------------...((....((((.....-.))))(((((.((((((...............(((((....)))))(.....).)))))))))))..))... ( -23.70, z-score = -1.61, R) >droSec1.super_5 3184641 93 - 5866729 -----------------GUAGGGGGGAAUGAGAUCUUU-CCCACCUGAUAGUUGCCGUCGAAUACUCCUCCGAAUGGAGACAUUCCACUACGCGGCAACAUCGGAGUCAUG -----------------((.((..(((......)))..-)).))(((((.((((((((.((....))....(((((....)))))......)))))))))))))....... ( -32.70, z-score = -2.07, R) >droSim1.chr2L 4911578 93 - 22036055 -----------------GUAGGGGGGAAUGAGAUCUUU-CCCACCUGAUAGUUGCCGUCGAAUACACCUCCGAAUGGAGACAUUCCACUACGCGGCAACAUCGGUGUCAUG -----------------...((..(((......)))..-))((((.(((.((((((((.((.......)).(((((....)))))......)))))))))))))))..... ( -33.70, z-score = -2.41, R) >droWil1.scaffold_180772 1202880 94 - 8906247 -----------------CUUAGGAUAGUCAUGAUUUCCACUUACCUGAUAGCUGCCUUCAAAUACACCACCAAAUGGCGUCAUGCCACUACGCGGCAGCAUUGGAGUCAUG -----------------...........(((((((.((((......)...((((((.......((.(((.....))).))...((......))))))))..)))))))))) ( -23.60, z-score = -1.26, R) >droPer1.super_1 5978523 74 + 10282868 ------------------------------------UU-CCCACCUGAUAGUUUCCAUCGAAUACCCCGCCGAAUGGCGUCAUUCCACUGCGCGGCAACAUAGGCGUCAUC ------------------------------------..-............................((((.....(((.((......)))))(....)...))))..... ( -12.70, z-score = 1.02, R) >anoGam1.chr3R 25762132 104 - 53272125 CCAUCGGGACGGCGAAUCGUCUGCGAGAUGAUGCUUUGGCCGAACAUACGCGGUCCCUGGAAUGCACCCUCAAACA-----AUCCCCCCUCGGGGAAACGCUG--CCCAAG .....(((.(((((.((((((.....))))))((((((((((........)))))....))).))...........-----.(((((....)))))..)))))--)))... ( -34.50, z-score = -0.87, R) >consensus __________________U_GGGGGAGAUGAGAUCUUU_CCCACCUGAUAGUUGCCAUCGAAUACACCUCCGAAUGGAGACAUUCCACUACGCGGCAACAUCGGAGUCAUG ............................................(((((.((((((...........((((....))))..............)))))))))))....... (-10.23 = -11.27 + 1.04)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:18:03 2011