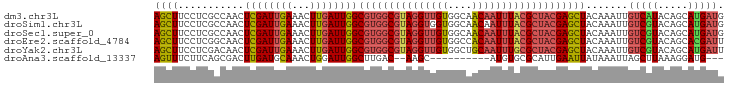
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 13,071,320 – 13,071,449 |
| Length | 129 |
| Max. P | 0.954671 |

| Location | 13,071,320 – 13,071,422 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 82.44 |
| Shannon entropy | 0.32999 |
| G+C content | 0.47988 |
| Mean single sequence MFE | -29.35 |
| Consensus MFE | -18.96 |
| Energy contribution | -20.70 |
| Covariance contribution | 1.74 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.565364 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
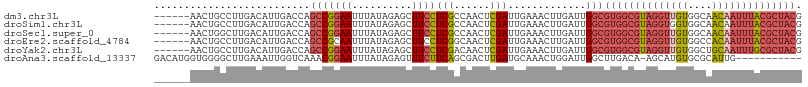
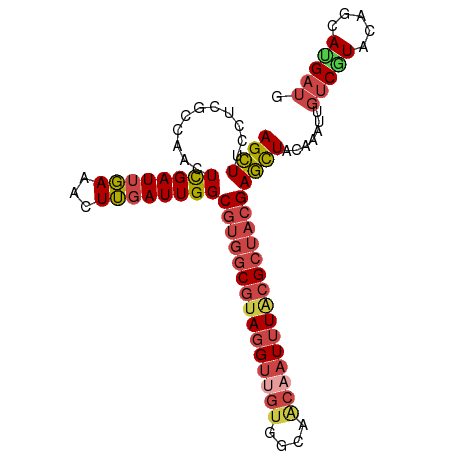
>dm3.chr3L 13071320 102 + 24543557 ------AACUGCCUUGACAUUGACCAGCCGGAAUUUAUAGAGCUUCCUCGCCAACUCGAUUGAAACUUGAUUGGCGUGGCGUAGGUUGUGGCAACAAUUUACGCUACG ------..(((.((.(........))).)))........(((....)))(((((.((((.......)))))))))((((((((((((((....)))))))))))))). ( -31.10, z-score = -2.34, R) >droSim1.chr3L 12462995 102 + 22553184 ------AACUGCCUUGACAUUGACCAGCCGGAAUUUAUAGAGCUUCCUCGCCAACUCGAUUGAAACUUGAUUGGCGUGGCGUAGGUGGUGGCAACAAUUUACGCUACG ------..(((.((.(........))).)))........(((....)))(((((.((((.......)))))))))(((((((((((..((....))))))))))))). ( -27.70, z-score = -0.91, R) >droSec1.super_0 5247169 102 + 21120651 ------AACUGGCUUGACAUUGACCAGCCGGAAUUUAUAGAGCUUCCUCGCCAACUCGAUUGAAACUUGAUUGGCGUGGCGUAGGUUGUGGCAACAAUUUACGCUACG ------..((((((.(........)))))))........(((....)))(((((.((((.......)))))))))((((((((((((((....)))))))))))))). ( -38.20, z-score = -4.43, R) >droEre2.scaffold_4784 13079460 102 + 25762168 ------AACUGCCUUGACAUUGACCAGCCGCAAUUUAUAGAGCUUCCUCGGCAACUCGAUUGAAACUUGAUUGGCGUGGCGUAGGUUGUGGCCACAAUUUACGCUACG ------...((((.....((((........)))).....(((....)))))))..(((((..(...)..)))))(((((((((((((((....))))))))))))))) ( -29.90, z-score = -1.59, R) >droYak2.chr3L 13154290 102 + 24197627 ------AACUGCCUUGACAUUGACCAGCCGGAAUUUAUAGAGCUUCCUCGACAACUCGAUUGAAACUUGAUUGGCGUGGCGUAGGUUGUGGCUGCAAUUUGCGCUACG ------....(((..........((....))........(((.(((.((((....))))..))).)))....)))((((((((((((((....)))))))))))))). ( -30.40, z-score = -1.63, R) >droAna3.scaffold_13337 10892472 96 - 23293914 GACAUGGUGGGGCUUGAAAUUGGUCAAACGGAAUUUAUAGAGUUUCUUCAGCGACUUGAUGCAAACUGGAUUGGCUUGACA-AGCAUGUGCGCAUUG----------- ...((((((.((((((.....((((((.(((....(((.(((((.(....).))))).)))....)))..))))))...))-))).).))).)))..----------- ( -18.80, z-score = 1.22, R) >consensus ______AACUGCCUUGACAUUGACCAGCCGGAAUUUAUAGAGCUUCCUCGCCAACUCGAUUGAAACUUGAUUGGCGUGGCGUAGGUUGUGGCAACAAUUUACGCUACG ..........................(((((((..........))))(((......))).............)))((((((((((((((....)))))))))))))). (-18.96 = -20.70 + 1.74)
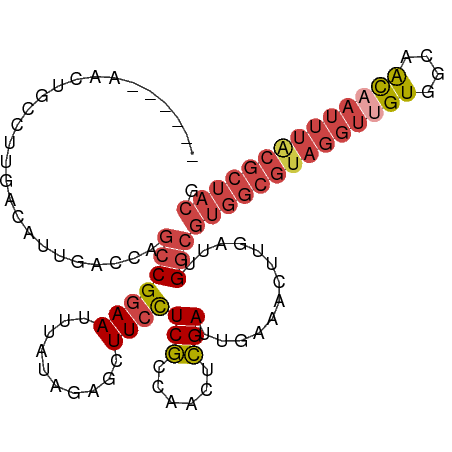
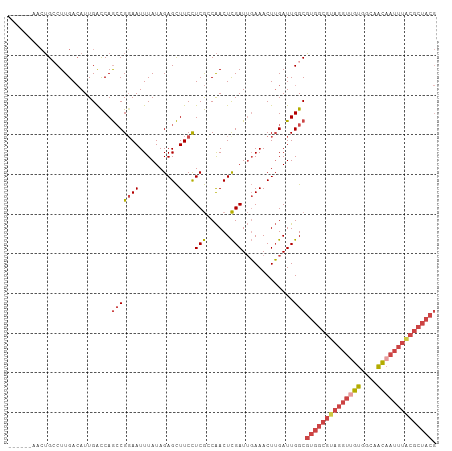
| Location | 13,071,354 – 13,071,449 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 82.88 |
| Shannon entropy | 0.33826 |
| G+C content | 0.46107 |
| Mean single sequence MFE | -30.13 |
| Consensus MFE | -19.93 |
| Energy contribution | -20.95 |
| Covariance contribution | 1.02 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.61 |
| SVM RNA-class probability | 0.954671 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 13071354 95 + 24543557 AGCUUCCUCGCCAACUCGAUUGAAACUUGAUUGGCGUGGCGUAGGUUGUGGCAACAAUUUACGCUACGAGCUACAAAUUGUCAUACAGCAUGAUG (((((....(((((.((((.......)))))))))((((((((((((((....))))))))))))))))))).......(((((.....))))). ( -35.10, z-score = -4.03, R) >droSim1.chr3L 12463029 95 + 22553184 AGCUUCCUCGCCAACUCGAUUGAAACUUGAUUGGCGUGGCGUAGGUGGUGGCAACAAUUUACGCUACGAGCUACAAAUUGUCGUACAGCAUGAUG (((((....(((((.((((.......)))))))))(((((((((((..((....)))))))))))))))))).......(((((.....))))). ( -31.30, z-score = -2.16, R) >droSec1.super_0 5247203 95 + 21120651 AGCUUCCUCGCCAACUCGAUUGAAACUUGAUUGGCGUGGCGUAGGUUGUGGCAACAAUUUACGCUACGAGCUACAAAUUGUCGUACAGCAUGAUG (((((....(((((.((((.......)))))))))((((((((((((((....))))))))))))))))))).......(((((.....))))). ( -34.70, z-score = -3.52, R) >droEre2.scaffold_4784 13079494 95 + 25762168 AGCUUCCUCGGCAACUCGAUUGAAACUUGAUUGGCGUGGCGUAGGUUGUGGCCACAAUUUACGCUACGAGCUACAAAUUGUCGUACAGCACGAUU .(((....((((((.(((((..(...)..)))))(((((((((((((((....))))))))))))))).........))))))...)))...... ( -33.50, z-score = -2.92, R) >droYak2.chr3L 13154324 95 + 24197627 AGCUUCCUCGACAACUCGAUUGAAACUUGAUUGGCGUGGCGUAGGUUGUGGCUGCAAUUUGCGCUACGAGCUACAAAUUGUCGUACAGCAUGAUU .(((....((((((.(((((..(...)..)))))(((((((((((((((....))))))))))))))).........))))))...)))...... ( -33.90, z-score = -2.78, R) >droAna3.scaffold_13337 10892512 80 - 23293914 AGUUUCUUCAGCGACUUGAUGCAAACUGGAUUGGCUUGAC--AAGC----------AUGUGCGCAUUGAAUUAUAAAUUAGCUUAAAGGAUG--- .(((..((.(((...(..((((..((((..(((......)--)).)----------).))..))))..)..((.....))))).))..))).--- ( -12.30, z-score = 1.53, R) >consensus AGCUUCCUCGCCAACUCGAUUGAAACUUGAUUGGCGUGGCGUAGGUUGUGGCAACAAUUUACGCUACGAGCUACAAAUUGUCGUACAGCAUGAUG ((((...........((((((((...))))))))(((((((((((((((....))))))))))))))))))).......(((((.....))))). (-19.93 = -20.95 + 1.02)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:22:44 2011