
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 13,057,581 – 13,057,713 |
| Length | 132 |
| Max. P | 0.925048 |

| Location | 13,057,581 – 13,057,691 |
|---|---|
| Length | 110 |
| Sequences | 12 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 79.02 |
| Shannon entropy | 0.44927 |
| G+C content | 0.41080 |
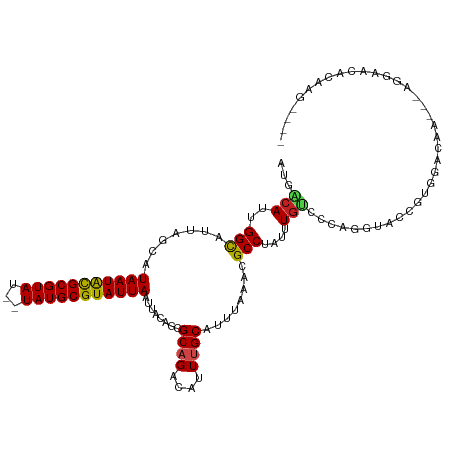
| Mean single sequence MFE | -26.18 |
| Consensus MFE | -18.39 |
| Energy contribution | -18.41 |
| Covariance contribution | 0.02 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.837933 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
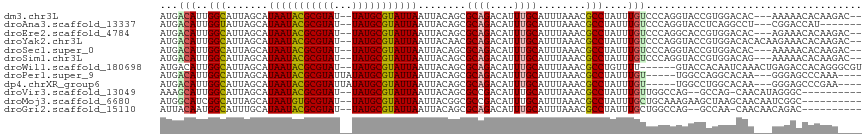
>dm3.chr3L 13057581 110 - 24543557 AUGACAUUGGCAUUAGCAUAAUACGCGUAU--UAUGCGUAUUAAUUACAGCGCAGACAUUUGCAUUUAAACGCCUAUUUGUCCCAGGUACCGUGGACAC---AAAAACACAAGAC-- ..((((..(((.......((((((((((..--.))))))))))........((((....))))........)))....))))((((....).)))....---.............-- ( -24.30, z-score = -1.19, R) >droAna3.scaffold_13337 10878917 105 + 23293914 AUGACAUUGGUAUUAGCAUAAUACGCGUAU--UAUGCGUAUUAAUUACAGCGCAGACAUUUGCAUUUAAACGCCUAUUUGUCCCAGGUACCUCAGGCCU---CGGACCAU------- ........(((.......((((((((((..--.))))))))))........((((....))))........))).....((((.((((.......))))---.))))...------- ( -25.70, z-score = -1.33, R) >droEre2.scaffold_4784 13065585 110 - 25762168 AUGACAUUGGCAUUAGCAUAAUACGCGUAU--UAUGCGUAUUAAUUACAGCGCAGACAUUUGCAUUUAAACGCCUAUUUGUCCCAGGCACCGUGGACAC---AGAAACACAAGAC-- ..((((..(((.......((((((((((..--.))))))))))........((((....))))........)))....))))((((....).)))....---.............-- ( -24.30, z-score = -1.12, R) >droYak2.chr3L 13139975 113 - 24197627 AUGACAUUGGCAUUAGCAUAAUACGCGUAU--UAUGCGUAUUAAUUACAACGCAGACAUUUGCAUUUAAACGCCUAUUUGUCCCAGGUACCGUGGACACACAAGAAACACAAGAC-- ..((((..(((.......((((((((((..--.))))))))))........((((....))))........)))....)))).........(((....)))..............-- ( -25.00, z-score = -1.73, R) >droSec1.super_0 5229213 110 - 21120651 AUGACAUUGGCAUUAGCAUAAUACGCGUAU--UAUGCGUAUUAAUUACAGCGCAGACAUUUGCAUUUAAACGCCUAUUUGUCCCAGGUACCGUGGACAC---AAAAACACAAGAC-- ..((((..(((.......((((((((((..--.))))))))))........((((....))))........)))....))))((((....).)))....---.............-- ( -24.30, z-score = -1.19, R) >droSim1.chr3L 12449311 110 - 22553184 AUGACAUUGGCAUUAGCAUAAUACGCGUAU--UAUGCGUAUUAAUUACAGCGCAGACAUUUGCAUUUAAACGCCUAUUUGUCCCAGGUACCGUGGACAG---AAAAACACAAGAC-- ........(((.......((((((((((..--.))))))))))........((((....))))........)))..(((((((..(....)..))))))---)............-- ( -26.40, z-score = -1.93, R) >droWil1.scaffold_180698 6539039 109 - 11422946 AUGACAUUGGCAUUAGCAUAAUACGCGUAU--UAUGCGUAUUAAUUACAGCGCAGACAUUUGCAUUUAAACGCCUGUUUU------GUACCACAAUCAAACUGAGACCACAGGGCGU .........((....((((((((....)))--)))))(((.....))).))((((....))))......(((((((..((------(.....))).))..(((......)))))))) ( -24.40, z-score = -0.58, R) >droPer1.super_9 3636132 105 - 3637205 AUGACAUUGGCAUUAGCAUAAUACGCGUAUUAUAUGCGUAUUAAUUACAGCGCAGACAUUUGCAUUUAAACGCCUAUUUGU-----UGGCCAGGCACAA---GGGAGCCCAAA---- .((...(((((..((((((((((((((((...)))))))))))........((((....))))...............)))-----))))))).))...---((....))...---- ( -25.60, z-score = -0.53, R) >dp4.chrXR_group6 5325862 105 - 13314419 AUGACAUUGGCAUUAGCAUAAUACGCGUAUUAUAUGCGUAUUAAUUACAGCGCAGACAUUUGCAUUUAAACGCCUAUUUGU-----UGGCCUGGCACAA---GGGAGCCCGAA---- ......(((((.......(((((((((((...)))))))))))....(((.((.((((...((........)).....)))-----).))))))).)))---((....))...---- ( -26.60, z-score = -0.50, R) >droVir3.scaffold_13049 12424078 102 - 25233164 AAAGCAUUGGCAUUAGCAUAAUACGCGUAU--UAUGCGUAUUAAUUACAGCGCCGACAUUUGCAUUUAAACGCCUAUUUGUUGGCCAG--GCCAG-CAACAUAGGGC---------- ...((((((((....((((((((....)))--)))))(((.....)))...)))))....))).........(((((((((((((...--)))))-))).)))))..---------- ( -35.20, z-score = -3.29, R) >droMoj3.scaffold_6680 9724450 105 + 24764193 AUGGCAUCGGCAUUAGCAUAAUGUGCGUAU--UAUGCGUAUUAAUUACGGCGCCGACAUUUGCAUUUAAACGCCUAUUUGCUGCAAAGAAGCUAAGCAACAAUCGGC---------- ...((((((((....((((((((....)))--)))))(((.....)))...)))))....)))........(((.(((((((((......))..))))..))).)))---------- ( -28.00, z-score = -0.08, R) >droGri2.scaffold_15110 23027671 102 - 24565398 AUUACAAUGGCAUUUGCAUAAUACGCGUAU--UAUGCGUAUUAAUUACAGCGCAGACAUUUGCAUUUAAACGCCUAUUUGCUGGCCAG--GCCAA-CAACAACAGAC---------- .......((((....((.((((((((((..--.))))))))))....((((((((....))))....(((......)))))))))...--)))).-...........---------- ( -24.30, z-score = -1.29, R) >consensus AUGACAUUGGCAUUAGCAUAAUACGCGUAU__UAUGCGUAUUAAUUACAGCGCAGACAUUUGCAUUUAAACGCCUAUUUGUCCCAGGUACCGUGGACAA___AGGAACACAAG____ ...(((..(((.......(((((((((((...)))))))))))........((((....))))........)))....))).................................... (-18.39 = -18.41 + 0.02)
| Location | 13,057,614 – 13,057,713 |
|---|---|
| Length | 99 |
| Sequences | 12 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 87.88 |
| Shannon entropy | 0.24188 |
| G+C content | 0.36579 |
| Mean single sequence MFE | -24.43 |
| Consensus MFE | -20.18 |
| Energy contribution | -20.15 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.83 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.925048 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
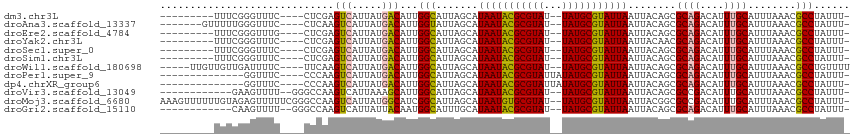
>dm3.chr3L 13057614 99 - 24543557 ---------UUUCGGGUUUC----CUCGAGUCAUUAUGACAUUGGCAUUAGCAUAAUACGCGUAU--UAUGCGUAUUAAUUACAGCGCAGACAUUUGCAUUUAAACGCCUAUUU- ---------..(((((....----)))))(((.....)))...(((.......((((((((((..--.))))))))))........((((....))))........))).....- ( -24.90, z-score = -2.34, R) >droAna3.scaffold_13337 10878945 101 + 23293914 -------GUUUUUGGGUUUC----CUCAAGUCAUUAUGACAUUGGUAUUAGCAUAAUACGCGUAU--UAUGCGUAUUAAUUACAGCGCAGACAUUUGCAUUUAAACGCCUAUUU- -------.....(((((...----.....(((.....)))((((((((..((((((((....)))--)))))))))))))......((((....))))........)))))...- ( -20.90, z-score = -1.08, R) >droEre2.scaffold_4784 13065618 99 - 25762168 ---------UUUCGGGUUUG----CUCGAGUCAUUAUGACAUUGGCAUUAGCAUAAUACGCGUAU--UAUGCGUAUUAAUUACAGCGCAGACAUUUGCAUUUAAACGCCUAUUU- ---------..(((((....----)))))(((.....)))...(((.......((((((((((..--.))))))))))........((((....))))........))).....- ( -26.10, z-score = -2.20, R) >droYak2.chr3L 13140011 99 - 24197627 ---------UUUCGGGUUUC----CUCGAGUCAUUAUGACAUUGGCAUUAGCAUAAUACGCGUAU--UAUGCGUAUUAAUUACAACGCAGACAUUUGCAUUUAAACGCCUAUUU- ---------..(((((....----)))))(((.....)))...(((.......((((((((((..--.))))))))))........((((....))))........))).....- ( -24.90, z-score = -2.79, R) >droSec1.super_0 5229246 99 - 21120651 ---------UUUCGGGUUUC----CUCGAGUCAUUAUGACAUUGGCAUUAGCAUAAUACGCGUAU--UAUGCGUAUUAAUUACAGCGCAGACAUUUGCAUUUAAACGCCUAUUU- ---------..(((((....----)))))(((.....)))...(((.......((((((((((..--.))))))))))........((((....))))........))).....- ( -24.90, z-score = -2.34, R) >droSim1.chr3L 12449344 99 - 22553184 ---------UUUCGGGUUUC----CUCGAGUCAUUAUGACAUUGGCAUUAGCAUAAUACGCGUAU--UAUGCGUAUUAAUUACAGCGCAGACAUUUGCAUUUAAACGCCUAUUU- ---------..(((((....----)))))(((.....)))...(((.......((((((((((..--.))))))))))........((((....))))........))).....- ( -24.90, z-score = -2.34, R) >droWil1.scaffold_180698 6539070 104 - 11422946 -----UUGUUGUUGAUUUUC----UUCAAGUCAUUAUGACAUUGGCAUUAGCAUAAUACGCGUAU--UAUGCGUAUUAAUUACAGCGCAGACAUUUGCAUUUAAACGCCUGUUUU -----.(((..(((((((..----...))))))..)..)))..(((.......((((((((((..--.))))))))))........((((....))))........)))...... ( -23.10, z-score = -1.06, R) >droPer1.super_9 3636158 96 - 3637205 --------------GGUUUC----CCCAAGUCAUUAUGACAUUGGCAUUAGCAUAAUACGCGUAUUAUAUGCGUAUUAAUUACAGCGCAGACAUUUGCAUUUAAACGCCUAUUU- --------------((....----.))..(((.....)))...(((.......(((((((((((...)))))))))))........((((....))))........))).....- ( -22.30, z-score = -2.12, R) >dp4.chrXR_group6 5325888 96 - 13314419 --------------GGUUUC----CCCAAGUCAUUAUGACAUUGGCAUUAGCAUAAUACGCGUAUUAUAUGCGUAUUAAUUACAGCGCAGACAUUUGCAUUUAAACGCCUAUUU- --------------((....----.))..(((.....)))...(((.......(((((((((((...)))))))))))........((((....))))........))).....- ( -22.30, z-score = -2.12, R) >droVir3.scaffold_13049 12424103 98 - 25233164 ------------GAAGUUUU--GGGCCAAGUCAUUAAAGCAUUGGCAUUAGCAUAAUACGCGUAU--UAUGCGUAUUAAUUACAGCGCCGACAUUUGCAUUUAAACGCCUAUUU- ------------.......(--((((...((..(((((((((((((....((((((((....)))--)))))(((.....)))...)))))....))).))))))))))))...- ( -25.30, z-score = -1.84, R) >droMoj3.scaffold_6680 9724478 112 + 24764193 AAAGUUUUUUGUAGAGUUUUUCGGGCCAAGUCAUUAUGGCAUCGGCAUUAGCAUAAUGUGCGUAU--UAUGCGUAUUAAUUACGGCGCCGACAUUUGCAUUUAAACGCCUAUUU- ...((((..((((((.........((((........)))).(((((....((((((((....)))--)))))(((.....)))...)))))..))))))...))))........- ( -30.10, z-score = -1.58, R) >droGri2.scaffold_15110 23027696 98 - 24565398 ------------CAAGUUUU--GGGCCAAGUCAUUAUUACAAUGGCAUUUGCAUAAUACGCGUAU--UAUGCGUAUUAAUUACAGCGCAGACAUUUGCAUUUAAACGCCUAUUU- ------------.......(--((((...((((((.....)))))).......((((((((((..--.))))))))))........((((....))))........)))))...- ( -23.50, z-score = -1.64, R) >consensus _________UUUCGGGUUUC____CUCAAGUCAUUAUGACAUUGGCAUUAGCAUAAUACGCGUAU__UAUGCGUAUUAAUUACAGCGCAGACAUUUGCAUUUAAACGCCUAUUU_ .............................(((.....)))...(((.......(((((((((((...)))))))))))........((((....))))........)))...... (-20.18 = -20.15 + -0.03)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:22:42 2011