| Sequence ID | dm3.chr3L |
|---|---|
| Location | 12,987,639 – 12,987,737 |
| Length | 98 |
| Max. P | 0.920059 |

| Location | 12,987,639 – 12,987,737 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 81.47 |
| Shannon entropy | 0.28583 |
| G+C content | 0.54249 |
| Mean single sequence MFE | -29.04 |
| Consensus MFE | -20.80 |
| Energy contribution | -21.84 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.837614 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
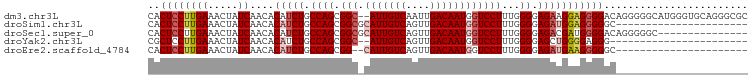
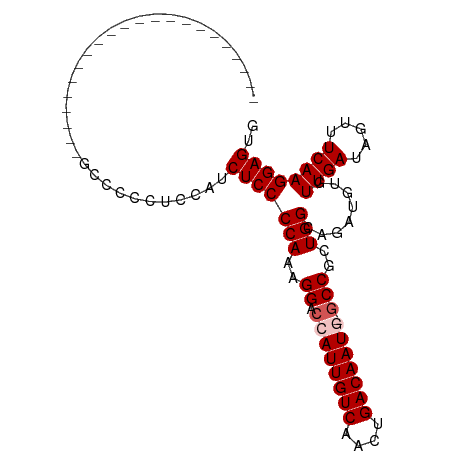
>dm3.chr3L 12987639 98 + 24543557 CACUCCUUGAAACUAUCAACACAUCUGCCAGCGGC--AUUGUCAAUUGACAAUGGUCCUUUGGGGAGAAGGAGGGGACAGGGGGCAUGGGUGCAGGGCGC ..(.(((((...((((...(...((((((.....(--((((((....))))))).((((((.....))))))..)).)))).)..))))...))))).). ( -30.20, z-score = -0.48, R) >droSim1.chr3L 12370499 78 + 22553184 CACUCCUUGAAACUAUCAACACAUCUGCCAGCGGCGCAUUGUCAGUUGACAAUGGUCCUUUGGGGAGAUGGAGGGGGC---------------------- ..((((((((.....))....(((((.((((.(((.(((((((....))))))))))))...)).)))))))))))..---------------------- ( -29.10, z-score = -2.45, R) >droSec1.super_0 5159303 85 + 21120651 CACUCCUUGAAACUAUCAACACAUCUGCCAGCGGCGCAUUGUCAGUUGACAAUGGUCCUUUGGGGAGACGAUGGGGACAGGGGGC--------------- ..(((((((...(((((..........((((.(((.(((((((....)))))))).)).))))(....))))))...))))))).--------------- ( -33.00, z-score = -3.02, R) >droYak2.chr3L 13065445 75 + 24197627 CGCUCCUUGAAACUAUCAACACAUCUGCCAGCGGC--AUUGUCAGUUGACAAUGGUCCUUUGGGGAGCUGGGGAGGG----------------------- (.((((((((.....))))........(((((..(--((((((....))))))).(((.....)))))))))))).)----------------------- ( -26.90, z-score = -1.73, R) >droEre2.scaffold_4784 12994102 76 + 25762168 CACUCCUUGAAACUAUCAACACAUCUGCCAGCGG--CAUUGUCAGUUGACAAUGGUCCUUUGGGGAGAUGAAGGGGGC---------------------- ..((((((((.....))....(((((.((...((--(((((((....)))))))..))....)).)))))))))))..---------------------- ( -26.00, z-score = -1.97, R) >consensus CACUCCUUGAAACUAUCAACACAUCUGCCAGCGGC_CAUUGUCAGUUGACAAUGGUCCUUUGGGGAGAUGGAGGGGGC______________________ ..((((((((.....))....(((((.((...((..(((((((....)))))))..))....)).)))))))))))........................ (-20.80 = -21.84 + 1.04)
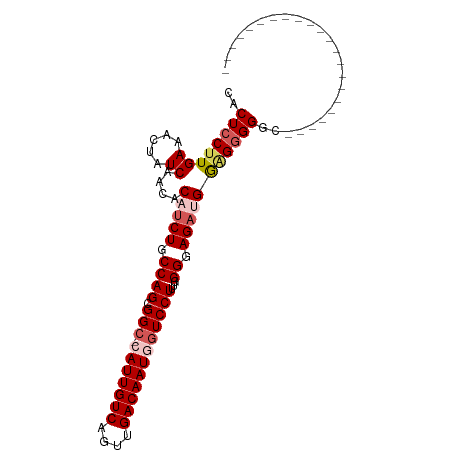
| Location | 12,987,639 – 12,987,737 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 81.47 |
| Shannon entropy | 0.28583 |
| G+C content | 0.54249 |
| Mean single sequence MFE | -23.90 |
| Consensus MFE | -18.14 |
| Energy contribution | -18.54 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.76 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.28 |
| SVM RNA-class probability | 0.920059 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 12987639 98 - 24543557 GCGCCCUGCACCCAUGCCCCCUGUCCCCUCCUUCUCCCCAAAGGACCAUUGUCAAUUGACAAU--GCCGCUGGCAGAUGUGUUGAUAGUUUCAAGGAGUG .((((((((((...((((..........(((((.......))))).(((((((....))))))--).....))))...))))(((.....)))))).))) ( -26.20, z-score = -1.78, R) >droSim1.chr3L 12370499 78 - 22553184 ----------------------GCCCCCUCCAUCUCCCCAAAGGACCAUUGUCAACUGACAAUGCGCCGCUGGCAGAUGUGUUGAUAGUUUCAAGGAGUG ----------------------.....((((((((..(((..((.((((((((....))))))).)))..))).))))...((((.....)))))))).. ( -24.10, z-score = -2.36, R) >droSec1.super_0 5159303 85 - 21120651 ---------------GCCCCCUGUCCCCAUCGUCUCCCCAAAGGACCAUUGUCAACUGACAAUGCGCCGCUGGCAGAUGUGUUGAUAGUUUCAAGGAGUG ---------------((.(((((((..((.(((((..(((..((.((((((((....))))))).)))..))).)))))))..)))))......)).)). ( -24.90, z-score = -2.08, R) >droYak2.chr3L 13065445 75 - 24197627 -----------------------CCCUCCCCAGCUCCCCAAAGGACCAUUGUCAACUGACAAU--GCCGCUGGCAGAUGUGUUGAUAGUUUCAAGGAGCG -----------------------..((((((((((((.....))).(((((((....))))))--)..)))))........((((.....)))))))).. ( -23.70, z-score = -2.46, R) >droEre2.scaffold_4784 12994102 76 - 25762168 ----------------------GCCCCCUUCAUCUCCCCAAAGGACCAUUGUCAACUGACAAUG--CCGCUGGCAGAUGUGUUGAUAGUUUCAAGGAGUG ----------------------((.((...(((((..(((..((..(((((((....)))))))--))..))).)))))..((((.....)))))).)). ( -20.60, z-score = -1.43, R) >consensus ______________________GCCCCCUCCAUCUCCCCAAAGGACCAUUGUCAACUGACAAUG_GCCGCUGGCAGAUGUGUUGAUAGUUUCAAGGAGUG .................................(((((((..((..(((((((....)))))))..))..)))........((((.....)))))))).. (-18.14 = -18.54 + 0.40)

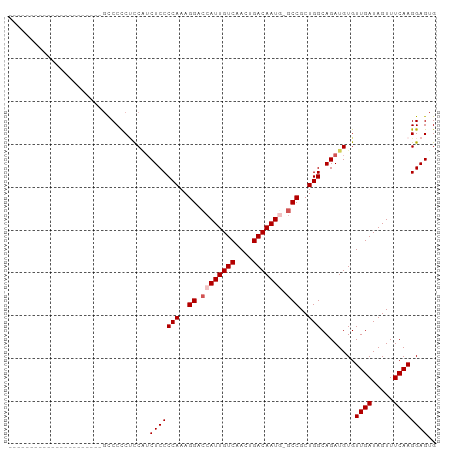
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:22:29 2011