| Sequence ID | dm3.chr3L |
|---|---|
| Location | 12,985,981 – 12,986,232 |
| Length | 251 |
| Max. P | 0.942495 |

| Location | 12,985,981 – 12,986,083 |
|---|---|
| Length | 102 |
| Sequences | 11 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 74.57 |
| Shannon entropy | 0.49591 |
| G+C content | 0.39280 |
| Mean single sequence MFE | -19.30 |
| Consensus MFE | -9.31 |
| Energy contribution | -9.07 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.852260 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
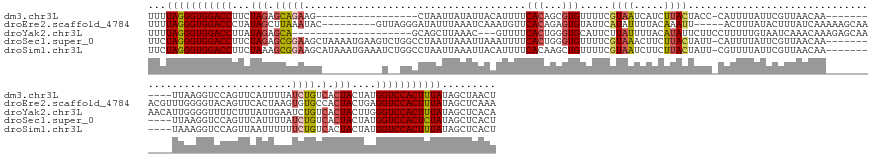
>dm3.chr3L 12985981 102 + 24543557 UUCGGAUCCCUAUUUAAUUAAGCACUUAAUUAUUUAAUGCGCUGAUUGCGCAAUAAUUCAAACGACCC---------------CUUGCUUGGCUCAGCUUAGCCAGAGUCCUUCGAA .(((((....((..(((((((....)))))))..)).(((((.....)))))...........(((..---------------((.(((.(((...))).))).)).))).))))). ( -23.30, z-score = -1.75, R) >droPer1.super_9 2647229 89 + 3637205 UCCGGCUCCUUAUUUAAUUAAGCACUUAAUUAUUUAAUGCGCUGAUUGUGCAAUAUUUCAAAGAACCC---------------CACCCUGGACCCCACAUCGUC------------- ...((.((((((..(((((((....)))))))..)))(((((.....)))))................---------------......))).)).........------------- ( -15.80, z-score = -1.94, R) >dp4.chrXR_group6 4358078 89 + 13314419 UCCGACUCCUUAUUUAAUUAAGCACUUAAUUAUUUAAUGCGCUGAUUGUGCAAUAUUUCAAAGAACCC---------------CACCCUGGACCCCACAUCGUC------------- ((((.....(((..(((((((....)))))))..)))(((((.....)))))................---------------.....))))............------------- ( -13.10, z-score = -1.93, R) >droAna3.scaffold_13337 6520964 117 - 23293914 UCCGUCUCCUUAUUUAAUUAAGCACUUAAUUAUUUAAUGCGCUGAUUGUGCAAUAUUUCAAAGGACCCAGCCCAGUACAUGACUCUGCUUAGCUCAGCUUAGCCAAUGUCCUUUGAG .........(((..(((((((....)))))))..)))(((((.....))))).....(((((((((..(((..(((.....)))..)))..(((......)))....))))))))). ( -26.90, z-score = -2.33, R) >droEre2.scaffold_4784 12992396 102 + 25762168 UCCUGCUUCCUAUUUAAUUAAGCACUUAAUUAUUUAAUGCGCUGAUUGCGCAAUAUUUCAAACGACUC---------------CAUGCUUAGCUCAACUUAGCCAGAGUCCUUCGAA ..........((..(((((((....)))))))..)).(((((.....)))))....(((.((.(((((---------------...(((.((.....)).)))..))))).)).))) ( -21.20, z-score = -2.44, R) >droYak2.chr3L 13063704 102 + 24197627 UCCUGCUCCCUAUUUAAUUAAGCACUUAAUUAUUUAAUGCGCUGAUUGCGCAAUAUUUCAACCGACUC---------------CUUGCUUAGCUCAGCUCAGCCAGAGUCCUUCGAU ..........((..(((((((....)))))))..)).(((((.....)))))...........(((((---------------...(((.(((...))).)))..)))))....... ( -22.90, z-score = -2.46, R) >droSec1.super_0 5157621 102 + 21120651 UUCGUAUCCCUAUUUAAUUAAGCACUUAAUUAUUUAAUGCGCUGAUUGCGCAAUAAUUCAAACGACCC---------------UUUGCUUGGCUCAGCUUAGCCAGAGUCCUUCGAA .((((.....((..(((((((....)))))))..)).(((((.....))))).........))))...---------------...((((((((......))))).)))........ ( -21.50, z-score = -1.68, R) >droSim1.chr3L 12368806 102 + 22553184 UUCGUAUCCCUAUUUAAUUAAGCACUUAAUUAUUUAAUGCGCUGAUUGCGCAAUAAUUCAAACGACCC---------------CUUGCUUGGCUCAGCUUAGCCAGAGUCCUUCGAA .((((.....((..(((((((....)))))))..)).(((((.....))))).........))))...---------------...((((((((......))))).)))........ ( -21.50, z-score = -1.75, R) >droVir3.scaffold_13049 2718683 100 - 25233164 UCCUGGUCACCAUUUAAUUAAGCACUUAAUUAUUUAAUGUG-UGAUUGCGCAAUAUUUCAAACGUAGC---------------CUUCG-CUCGUCUACUUAAUGACAACUCAUCCAA ....(((((((((((((((((....)))))))...)))).)-)))))................(.(((---------------....)-)))(((........)))........... ( -14.80, z-score = -0.97, R) >droMoj3.scaffold_6680 7636824 102 - 24764193 UCCUGGUCUCCGUUUAAUUAAGUACUUAAUUAUUUAAUGCGCUGAUUGUGCAAUAUUUCAAACGUUGC---------------CUUCAACUCGUCUGCUUAAUGACAACUCAUCCAA ....(((...(((((.((((((((......))))))))((((.....))))........)))))..))---------------)........(((........)))........... ( -17.50, z-score = -2.07, R) >droGri2.scaffold_15110 21026165 101 + 24565398 UCCUGGUCGCCAUUUAAUUAAGCACUUAAUUAUUUAAUGCGCUGAUUGUGCAAUAUUUCAAACGUAUC---------------CUUUG-CUCCUCGACUUAAUGACAACUCAUCCAA ...(((..........(((((((..............(((((.....)))))...........(((..---------------...))-).....).))))))..........))). ( -13.85, z-score = -0.71, R) >consensus UCCGGCUCCCUAUUUAAUUAAGCACUUAAUUAUUUAAUGCGCUGAUUGCGCAAUAUUUCAAACGACCC_______________CUUGCUUGGCUCAGCUUAGCCAGAGUCCUUCGAA ..............(((((((....))))))).....(((((.....)))))................................................................. ( -9.31 = -9.07 + -0.24)

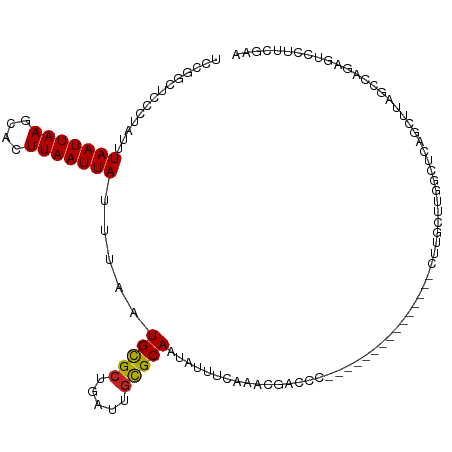
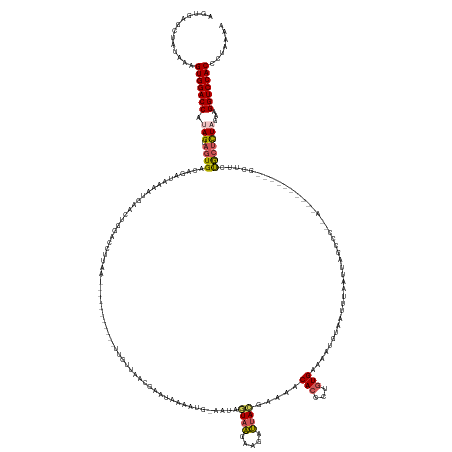
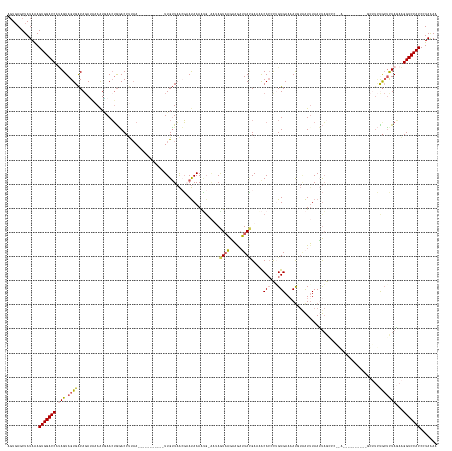
| Location | 12,986,083 – 12,986,232 |
|---|---|
| Length | 149 |
| Sequences | 5 |
| Columns | 178 |
| Reading direction | forward |
| Mean pairwise identity | 67.37 |
| Shannon entropy | 0.55948 |
| G+C content | 0.34634 |
| Mean single sequence MFE | -33.76 |
| Consensus MFE | -14.14 |
| Energy contribution | -14.26 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.748693 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 12986083 149 + 24543557 AGUUAGCUAUAAAGUGGACCAUAGUAGUGACAGAUAAAAUGAACUGGACCUUAA-----------UUGUUAACGAAUAAAAUG-GGUAGUAAGAUGAUUACGAAAACACGCUGUGAAAAUGUAAUAUAAUUAG-----------------CUUCUGCUCUAGAAGGUCCACCCUAAAA .............((((((((((((.(((.(((..........))).((((...-----------(((((....)))))...)-))).((((.....)))).....)))))))))..................-----------------((((((...))))))))))))....... ( -29.30, z-score = -0.91, R) >droEre2.scaffold_4784 12992498 164 + 25762168 UUUGAGCUAUAAAGUGGACCUCAGUAGUGGCACACUUAGUGAACUGUACCCCAAACGUUUGCUUUUUGAUAAAGUAUAAAGU-----AAUUUGUAAAAUAUGAAUACACUCUGUGAACAUUUGAUUUAAAUAUCCCUAAC---------GUAUUUAAGCUAUAGGGUCCACCCUAAAA .............((((((((((.(((((...))).)).))).(((((...((((.(((..(...((.(((...((((((..-----..))))))...))).))........)..))).)))).(((((((((.......---------))))))))).))))))))))))....... ( -33.30, z-score = -0.85, R) >droYak2.chr3L 13063806 155 + 24197627 UGUGAGCUAUAAAGUGGACCCAAGUAGUGACAGAUUCAAUAAAGAAAACCCCAAUGUUUUGCUCUUUGUUUGAUUACAAAAAGGAAGAAUAUGUAAAAUAAGAAUGCACCCAGUGAAAAC---GUUUAAGCUGC--------------------UGCUCUAUAAGGUCCACCCUAAAA .............(((((((...((((.(.(((.....................(((((((((((((.(((........))).)))))....)))))))).....((.....((....))---......))..)--------------------))).))))..)))))))....... ( -31.00, z-score = -1.45, R) >droSec1.super_0 5157723 166 + 21120651 AGUGAGCUAUAGAGUGGACCAUAGUAGUGACAGAUAAAAUGAACUGGACCUUAA-----------UUGUUAACGAAUAAAAUG-AAUAGUAAGAAGUUUACGAAAACACCCAGUGAAAAUUUAAUUUAAUUAGGCCAGACUUCAUUUUAGCUUCCGCUCUAGAAGGUCCACCCUAGAA .........(((.(((((((.(((.((((..((.(((((((((((((.(((.((-----------(((...............-....(((((...))))).....(((...)))...........))))))))))))..))))))))).))..)))))))...))))))).)))... ( -41.80, z-score = -3.60, R) >droSim1.chr3L 12368908 166 + 22553184 AGUGAGCUAUAAAGUGGACCAUAGUAGUGACAGAAAAAAUUAACUGGACCUUUA-----------UUGUUAACGAAUAAAACG-AAUAGUAAGAAGAUUACGAAAACAGCUUGUGAAAAUGUAAUUUAAUUAGGCCAGAUUUCAUUUAUGCUUCCGCUUUAGAAGGUCCACCCUAGAA .(((..((.(((((((((.(((((((((..........)))).((((.((((((-----------(((((..((.......))-))))))))..((((((((...((.....)).....))))))))....))))))).......)))))..)))))))))...))..)))....... ( -33.40, z-score = -1.35, R) >consensus AGUGAGCUAUAAAGUGGACCAUAGUAGUGACAGAUAAAAUGAACUGGACCUUAA___________UUGUUAACGAAUAAAAUG_AAUAGUAAGAAGAUUACGAAAACACCCUGUGAAAAUGUAAUUUAAUUAGCCC__A__________GCUUCUGCUCUAGAAGGUCCACCCUAAAA .............(((((((.(((.((((...........................................................((((.....)))).....(((...))).......................................)))))))...)))))))....... (-14.14 = -14.26 + 0.12)
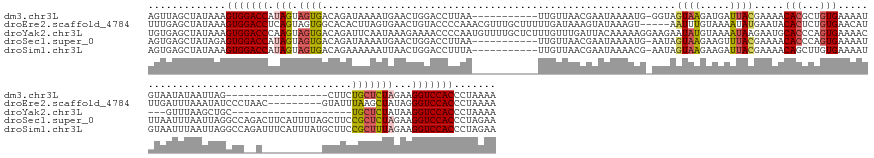
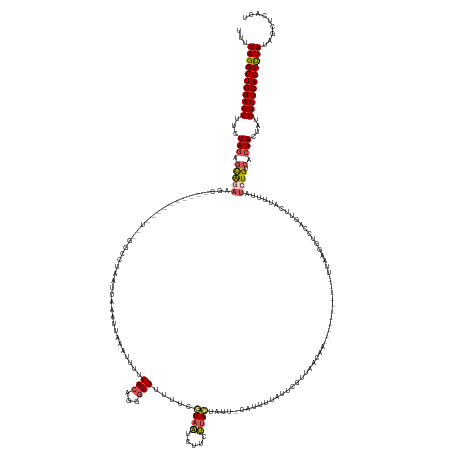
| Location | 12,986,083 – 12,986,232 |
|---|---|
| Length | 149 |
| Sequences | 5 |
| Columns | 178 |
| Reading direction | reverse |
| Mean pairwise identity | 67.37 |
| Shannon entropy | 0.55948 |
| G+C content | 0.34634 |
| Mean single sequence MFE | -34.54 |
| Consensus MFE | -16.18 |
| Energy contribution | -16.50 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.48 |
| SVM RNA-class probability | 0.942495 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 12986083 149 - 24543557 UUUUAGGGUGGACCUUCUAGAGCAGAAG-----------------CUAAUUAUAUUACAUUUUCACAGCGUGUUUUCGUAAUCAUCUUACUACC-CAUUUUAUUCGUUAACAA-----------UUAAGGUCCAGUUCAUUUUAUCUGUCACUACUAUGGUCCACUUUAUAGCUAACU ...(((((((((((...(((.(((((.(-----------------((...................))).((((..((................-.........))..)))).-----------....................)))))..)))....)))))))))))......... ( -25.52, z-score = -0.78, R) >droEre2.scaffold_4784 12992498 164 - 25762168 UUUUAGGGUGGACCCUAUAGCUUAAAUAC---------GUUAGGGAUAUUUAAAUCAAAUGUUCACAGAGUGUAUUCAUAUUUUACAAAUU-----ACUUUAUACUUUAUCAAAAAGCAAACGUUUGGGGUACAGUUCACUAAGUGUGCCACUACUGAGGUCCACUUUAUAGCUCAAA ...((((((((((((..(((((.....((---------(((..((((((((.....))))))))..((((((((.....(((.....))).-----....))))))))...........)))))..))((((((.((....)).)))))).)))..).)))))))))))......... ( -39.70, z-score = -2.28, R) >droYak2.chr3L 13063806 155 - 24197627 UUUUAGGGUGGACCUUAUAGAGCA--------------------GCAGCUUAAAC---GUUUUCACUGGGUGCAUUCUUAUUUUACAUAUUCUUCCUUUUUGUAAUCAAACAAAGAGCAAAACAUUGGGGUUUUCUUUAUUGAAUCUGUCACUACUUGGGUCCACUUUAUAGCUCACA ...((((((((((((..(((....--------------------(((.((.....---.........)).)))...........(((.((((...((((((((......)))))))).(((((......))))).......)))).)))..)))...))))))))))))......... ( -34.34, z-score = -1.36, R) >droSec1.super_0 5157723 166 - 21120651 UUCUAGGGUGGACCUUCUAGAGCGGAAGCUAAAAUGAAGUCUGGCCUAAUUAAAUUAAAUUUUCACUGGGUGUUUUCGUAAACUUCUUACUAUU-CAUUUUAUUCGUUAACAA-----------UUAAGGUCCAGUUCAUUUUAUCUGUCACUACUAUGGUCCACUCUAUAGCUCACU ...(((((((((((...(((((.(..((.(((((((((..(((((((((((...((((.........(((((.....((((.....))))....-)))))......)))).))-----------)).))).))))))))))))).))..).)).))).)))))))))))......... ( -42.06, z-score = -3.33, R) >droSim1.chr3L 12368908 166 - 22553184 UUCUAGGGUGGACCUUCUAAAGCGGAAGCAUAAAUGAAAUCUGGCCUAAUUAAAUUACAUUUUCACAAGCUGUUUUCGUAAUCUUCUUACUAUU-CGUUUUAUUCGUUAACAA-----------UAAAGGUCCAGUUAAUUUUUUCUGUCACUACUAUGGUCCACUUUAUAGCUCACU ...(((((((((((.......(((((((...((((.....(((((((..............................((((.....))))....-.(((.........)))..-----------...))).))))...))))))))))).........)))))))))))......... ( -31.09, z-score = -1.43, R) >consensus UUUUAGGGUGGACCUUCUAGAGCAGAAGC__________U__GGCCUAAUUAAAUUAAAUUUUCACAGGGUGUUUUCGUAAUCUUCUUACUAUU_CAUUUUAUUCGUUAACAA___________UUAAGGUCCAGUUCAUUUUAUCUGUCACUACUAUGGUCCACUUUAUAGCUCACU ...(((((((((((...(((.(((((.....................................(((...))).....((((.....))))......................................................)))).).)))....)))))))))))......... (-16.18 = -16.50 + 0.32)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:22:28 2011