| Sequence ID | dm3.chr3L |
|---|---|
| Location | 12,970,085 – 12,970,178 |
| Length | 93 |
| Max. P | 0.658161 |

| Location | 12,970,085 – 12,970,178 |
|---|---|
| Length | 93 |
| Sequences | 8 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 77.67 |
| Shannon entropy | 0.41136 |
| G+C content | 0.54469 |
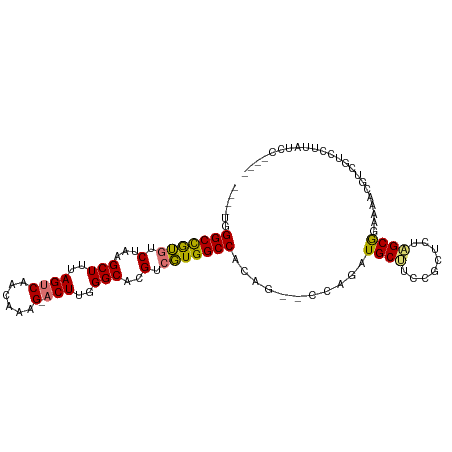
| Mean single sequence MFE | -31.81 |
| Consensus MFE | -16.47 |
| Energy contribution | -16.06 |
| Covariance contribution | -0.41 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.658161 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
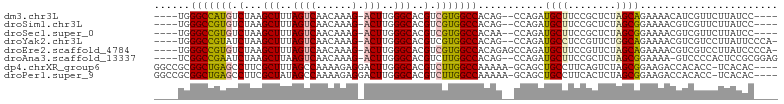
>dm3.chr3L 12970085 93 + 24543557 ----UGGGCCAUGUCUAAGCUUUAGUCAACAAAG-ACUUGGGCACGUCGUGGCCACAG--CCAGAUGCUUCCGCUCUAGCAGAAAACAUCGUUCUUAUCC---- ----(((((((((.(...(((..((((......)-)))..)))..).))))))).)).--...((((.(((.((....)).)))..))))..........---- ( -29.40, z-score = -2.28, R) >droSim1.chr3L 12352839 93 + 22553184 ----UGGGCCGUGUCUAAGCUUUAGUCAACAAAG-ACUUGGGCACGUCGUGGCCACAG--CCAGAUGCUUCCGCUCUAGCGGAAAACGUCGUUCUUAUCC---- ----.((((((((((((((((((.(....)))))-.))))))))))...((((....)--)))((((.((((((....))))))..))))))))......---- ( -34.80, z-score = -2.94, R) >droSec1.super_0 5141710 93 + 21120651 ----UGGGCCGUGUCUAAGCUUUAGUCAACAAAG-ACUUGGGCACGUCGUGGCCACAA--CCAGAUGCUUCCGCUCUAGCGGAAAACGUCGUUCUUAUCC---- ----(((((((((((((((((((.(....)))))-.))))))))).....)))).)).--...((((.((((((....))))))..))))..........---- ( -32.90, z-score = -2.76, R) >droYak2.chr3L 13047724 96 + 24197627 ----UGGGCCGUAUCUAAGCUUUAGUCAACAAAG-ACUUGGGCACGUCGUGGCCACAG--CCAGAUGCCUCCGUUCUGGCAGAAAACGUCGUCCUUAUUCCCA- ----.(((..........(((..((((......)-)))..)))(((.((((((....)--))...((((........))))....))).))).......))).- ( -28.80, z-score = -1.19, R) >droEre2.scaffold_4784 12976764 98 + 25762168 ----UGGGCCGUGUCUAAGCUUUAGUCAACAAAG-ACUUGGGCACGUCGUGGCCACAGAGCCAGAUGCUUCCGUUCUAGCAGAAAACGUCGUCCUUAUCCCCA- ----.((((.(((((((((((((.(....)))))-.)))))))))(.((((((......)))...((((........))))....))).))))).........- ( -30.10, z-score = -1.43, R) >droAna3.scaffold_13337 6504790 96 - 23293914 ----UCGGCCGAAUCUAAGCUUAAGUCAACAAAG-ACUUGGGCACGUCUUGGCCACAG--CCAGAUGCUUCCGCUCUAGCGGAAAA-GUCCCCACUCCGCGGAG ----..((((((..(...(((((((((......)-))))))))..)..))))))....--...(((..((((((....))))))..-)))....(((....))) ( -36.10, z-score = -3.06, R) >dp4.chrXR_group6 4341423 98 + 13314419 GGCCGCGGCUGAGCCUUCGCUUUAGCCAAAAGAGGACUUGGGCACGUCUUGGCCAAAAA-GCAGCUGCCUUCAGUCUAGCGGAAGACCACACC-UCACAC---- ((((((((((((((....)).))))))......(((((.(((((.((....((......-)).)))))))..))))).))))....)).....-......---- ( -31.80, z-score = -0.27, R) >droPer1.super_9 2630690 98 + 3637205 GGCCGCGGCUGAGCCUUCGCUAUAGCCAAAAGAGGACUUGGGCACGUCUUGGCCAAAAA-GCAGCUGCCUUCACUCUAGCGGAAGACCACACC-UCACAC---- ((((((((((((((....))).)))))...((((.....(((((.((....((......-)).)))))))...)))).))))....)).....-......---- ( -30.60, z-score = -0.45, R) >consensus ____UGGGCCGUGUCUAAGCUUUAGUCAACAAAG_ACUUGGGCACGUCGUGGCCACAG__CCAGAUGCUUCCGCUCUAGCGGAAAACGUCGUCCUUAUCC____ ......(((((((.(...(((..((((......).)))..)))..).)))))))...........((((........))))....................... (-16.47 = -16.06 + -0.41)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:22:25 2011