| Sequence ID | dm3.chr3L |
|---|---|
| Location | 12,968,650 – 12,968,745 |
| Length | 95 |
| Max. P | 0.866998 |

| Location | 12,968,650 – 12,968,745 |
|---|---|
| Length | 95 |
| Sequences | 8 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 88.79 |
| Shannon entropy | 0.20290 |
| G+C content | 0.33840 |
| Mean single sequence MFE | -8.38 |
| Consensus MFE | -7.71 |
| Energy contribution | -7.52 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.14 |
| Structure conservation index | 0.92 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.700097 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 12968650 95 + 24543557 GCCACCAUUUUGCUCAAUUUAUUUCCAACAUCAAGCUCUC----UCGUUUCCUUUUUUUUUUUAAUUU-AACACAAAAUUUUCGUGUAAUGUUUGGCUUU ((((.((((.......................((((....----..))))..................-.((((.........))))))))..))))... ( -8.30, z-score = -0.77, R) >droSim1.chr3L 12351371 91 + 22553184 GCCACCAUUUUGCUCAAUUUAUUUCCAACAUCAAGCUCUC----UCGUUCCCUUUUUUU----AAUUU-AACACACAAUUUUCGUGUAAUGUUUGGCUUU ((((.((((..(((...................)))....----...............----.....-...((((.......))))))))..))))... ( -8.61, z-score = -1.27, R) >droSec1.super_0 5140261 91 + 21120651 GCCACCAUUUUGCUCAAUUUAUUUCCAACAUCAAGCUCUC----UCGUUCCCUUUUUUU----AAUUU-AACACACAAUUUUCGUGUAAUGUUUGGCUUU ((((.((((..(((...................)))....----...............----.....-...((((.......))))))))..))))... ( -8.61, z-score = -1.27, R) >droYak2.chr3L 13046303 90 + 24197627 GCCACCAUUUUGCUCAAUUUAUUUCCAACAUCAAGCUCCC----UCGUUCCCUUUUUU-----AAUUU-AACACACAAUUUUCGUGUAAUGUUUGGCUUU ((((.((((..(((...................)))....----..............-----.....-...((((.......))))))))..))))... ( -8.61, z-score = -1.18, R) >droEre2.scaffold_4784 12975245 90 + 25762168 GCCACCAUUUUGCUCAAUUUAUUUCCAACAUCAAGCUCCC----UCGUUCCCUUUUUU-----AAUUU-AACACACAAUUUUCGUGUAAUGUUUGGCUUU ((((.((((..(((...................)))....----..............-----.....-...((((.......))))))))..))))... ( -8.61, z-score = -1.18, R) >droAna3.scaffold_13337 6503340 88 - 23293914 GCCACCAUUUUGCUCAAUUUAUUUCCAACAUCAAGCUCCC----A---UUCCUUUUUUU----AAUUU-UACACACAAUUUUCGUGUAAUGUUUGGCUUU ((((.......(((...................)))....----.---...........----(((.(-(((((.........)))))).)))))))... ( -9.51, z-score = -1.85, R) >dp4.chrXR_group6 4339785 100 + 13314419 GCCACCAUUUUGCUCAAUUUAUUUCCAACAUCAAGUCCUCGCUCACUCCUCUUUUCUUUUUUUAAUUUCAACACAAAAUUUUCGUGUAAUGUUUGGCUUU ((((.((((........................(((....)))...........................((((.........))))))))..))))... ( -7.40, z-score = -0.80, R) >droPer1.super_9 2629056 100 + 3637205 GCCACCAUUUUGCUCAAUUUAUUUCCAACAUCAAGUCCUCGCUCACUCCUCUUUUCUUUUUUUAAUUUCAACACAAAAUUUUCGUGUAAUGUUUGGCUUU ((((.((((........................(((....)))...........................((((.........))))))))..))))... ( -7.40, z-score = -0.80, R) >consensus GCCACCAUUUUGCUCAAUUUAUUUCCAACAUCAAGCUCUC____UCGUUCCCUUUUUUU____AAUUU_AACACACAAUUUUCGUGUAAUGUUUGGCUUU ((((.((((..(((...................)))..................................((((.........))))))))..))))... ( -7.71 = -7.52 + -0.19)

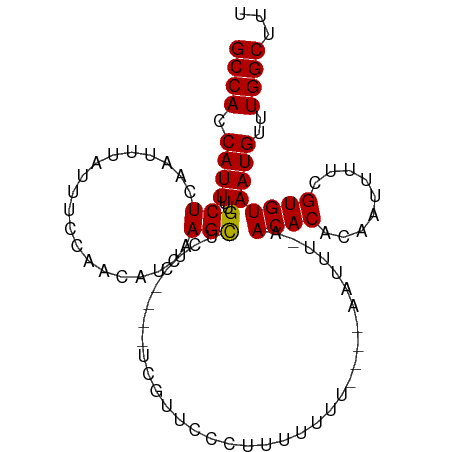
| Location | 12,968,650 – 12,968,745 |
|---|---|
| Length | 95 |
| Sequences | 8 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 88.79 |
| Shannon entropy | 0.20290 |
| G+C content | 0.33840 |
| Mean single sequence MFE | -16.11 |
| Consensus MFE | -13.77 |
| Energy contribution | -14.02 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.85 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.866998 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 12968650 95 - 24543557 AAAGCCAAACAUUACACGAAAAUUUUGUGUU-AAAUUAAAAAAAAAAAGGAAACGA----GAGAGCUUGAUGUUGGAAAUAAAUUGAGCAAAAUGGUGGC ...((((..((((((((((.....)))))).-................(....)..----....(((..((..(....)...))..)))..)))).)))) ( -18.30, z-score = -2.92, R) >droSim1.chr3L 12351371 91 - 22553184 AAAGCCAAACAUUACACGAAAAUUGUGUGUU-AAAUU----AAAAAAAGGGAACGA----GAGAGCUUGAUGUUGGAAAUAAAUUGAGCAAAAUGGUGGC ...((((..((((((((((...))))))...-.....----...............----....(((..((..(....)...))..)))..)))).)))) ( -16.70, z-score = -2.17, R) >droSec1.super_0 5140261 91 - 21120651 AAAGCCAAACAUUACACGAAAAUUGUGUGUU-AAAUU----AAAAAAAGGGAACGA----GAGAGCUUGAUGUUGGAAAUAAAUUGAGCAAAAUGGUGGC ...((((..((((((((((...))))))...-.....----...............----....(((..((..(....)...))..)))..)))).)))) ( -16.70, z-score = -2.17, R) >droYak2.chr3L 13046303 90 - 24197627 AAAGCCAAACAUUACACGAAAAUUGUGUGUU-AAAUU-----AAAAAAGGGAACGA----GGGAGCUUGAUGUUGGAAAUAAAUUGAGCAAAAUGGUGGC ...((((..((((((((((...))))))...-.....-----..............----....(((..((..(....)...))..)))..)))).)))) ( -16.70, z-score = -1.94, R) >droEre2.scaffold_4784 12975245 90 - 25762168 AAAGCCAAACAUUACACGAAAAUUGUGUGUU-AAAUU-----AAAAAAGGGAACGA----GGGAGCUUGAUGUUGGAAAUAAAUUGAGCAAAAUGGUGGC ...((((..((((((((((...))))))...-.....-----..............----....(((..((..(....)...))..)))..)))).)))) ( -16.70, z-score = -1.94, R) >droAna3.scaffold_13337 6503340 88 + 23293914 AAAGCCAAACAUUACACGAAAAUUGUGUGUA-AAAUU----AAAAAAAGGAA---U----GGGAGCUUGAUGUUGGAAAUAAAUUGAGCAAAAUGGUGGC ...((((....(((((((.......))))))-)....----...........---.----....(((..((..(....)...))..))).......)))) ( -16.80, z-score = -2.53, R) >dp4.chrXR_group6 4339785 100 - 13314419 AAAGCCAAACAUUACACGAAAAUUUUGUGUUGAAAUUAAAAAAAGAAAAGAGGAGUGAGCGAGGACUUGAUGUUGGAAAUAAAUUGAGCAAAAUGGUGGC ...((((..((((((((((.....))))))......................((((........))))..((((.((......)).)))).)))).)))) ( -13.50, z-score = -0.38, R) >droPer1.super_9 2629056 100 - 3637205 AAAGCCAAACAUUACACGAAAAUUUUGUGUUGAAAUUAAAAAAAGAAAAGAGGAGUGAGCGAGGACUUGAUGUUGGAAAUAAAUUGAGCAAAAUGGUGGC ...((((..((((((((((.....))))))......................((((........))))..((((.((......)).)))).)))).)))) ( -13.50, z-score = -0.38, R) >consensus AAAGCCAAACAUUACACGAAAAUUGUGUGUU_AAAUU____AAAAAAAGGAAACGA____GAGAGCUUGAUGUUGGAAAUAAAUUGAGCAAAAUGGUGGC ...((((..(((((((((.......)))))..................................(((..((..(....)...))..)))..)))).)))) (-13.77 = -14.02 + 0.25)



Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:22:24 2011