| Sequence ID | dm3.chr3L |
|---|---|
| Location | 12,924,926 – 12,925,031 |
| Length | 105 |
| Max. P | 0.993095 |

| Location | 12,924,926 – 12,925,031 |
|---|---|
| Length | 105 |
| Sequences | 10 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 84.56 |
| Shannon entropy | 0.31795 |
| G+C content | 0.41119 |
| Mean single sequence MFE | -25.87 |
| Consensus MFE | -20.96 |
| Energy contribution | -20.97 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.59 |
| SVM RNA-class probability | 0.993095 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
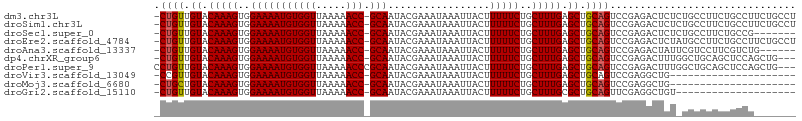
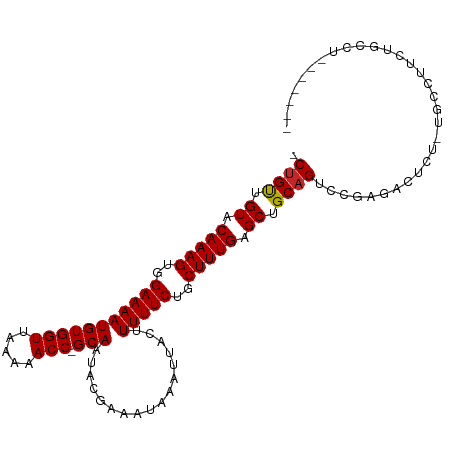
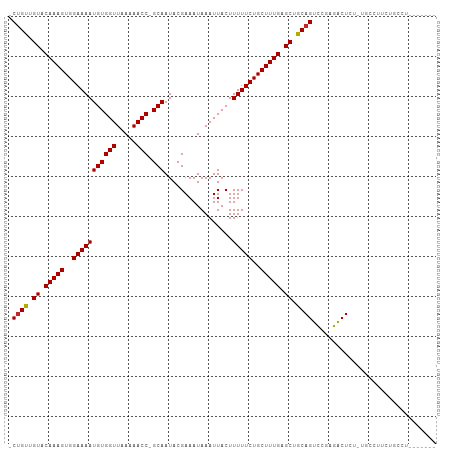
>dm3.chr3L 12924926 105 + 24543557 -CUGUUGUACAAAGUGGAAAAUGUGGUUAAAAACC-GCAAUACGAAAUAAAUUACUUUUUCUGCUUUGAGCUGCAGUCCGAGACUCUCUGCCUUCUGCCUUCUGCCU -((((.((.(((((..(((((((((((.....)))-))).................)))))..))))).)).))))...(((...)))................... ( -23.12, z-score = -1.60, R) >droSim1.chr3L 12304196 105 + 22553184 -CUGUUGUACAAAGUGGAAAAUGUGGUUAAAAACC-GCAAUACGAAAUAAAUUACUUUUUCUGCUUUGAGCUGCAGUCCGAGACUCUCUGCCUUCUGCCUUCUGCCU -((((.((.(((((..(((((((((((.....)))-))).................)))))..))))).)).))))...(((...)))................... ( -23.12, z-score = -1.60, R) >droSec1.super_0 5100142 98 + 21120651 -CUGUUGUACAAAGUGGAAAAUGUGGUUAAAAACC-GCAAUACGAAAUAAAUUACUUUUUCUGCUUUGAGCUGCAGUCCGAGACUCUCUGCCUUCUGCCG------- -((((.((.(((((..(((((((((((.....)))-))).................)))))..))))).)).))))...(((...)))............------- ( -23.12, z-score = -2.00, R) >droEre2.scaffold_4784 12934299 105 + 25762168 -CUGUUGUACAAAGUGGAAAAUGUGGUUAAAAACC-GCAAUACGAAAUAAAUUACUUUUUCUGCUUUGAGCUGCAGUCCGAGACUCUAUGCCUUCUGCCUUCUGCCU -((((.((.(((((..(((((((((((.....)))-))).................)))))..))))).)).))))............................... ( -22.82, z-score = -1.57, R) >droAna3.scaffold_13337 6468657 99 - 23293914 -CUGUUGUACAAAGUGGAAAAUGUGGUUAAAAACC-GCAAUACGAAAUAAAUUACUUUUUCUGCUUUGAGCUGCAGUCCGAGACUAUUCGUCCUUCGUCUG------ -((((.((.(((((..(((((((((((.....)))-))).................)))))..))))).)).))))..((((((.....)))..)))....------ ( -25.62, z-score = -2.93, R) >dp4.chrXR_group6 4299473 102 + 13314419 -CUGUUGUACAAAGUGGAAAAUGUGGUUAAAAACC-GCAAUACGAAAUAAAUUACUUUUUCUGCUUUGAGCUGCAGUCCGAGACUUUGGCUGCAGCUCCAGCUG--- -..((((...((((..(((((((((((.....)))-))).................)))))..))))(((((((((.((((....)))))))))))))))))..--- ( -36.62, z-score = -4.67, R) >droPer1.super_9 2588153 104 + 3637205 CCUGUUGUACAAAGUGGAAAAUGUGGUUAAAAACCCGCAAUACGAAAUAAAUUACUUUUUCUGCUUUGAGCUGCAGUCCGAGACUUUGGCUGCAGCUCCAGCUG--- ...((((...((((..(((((.((((((.......((.....)).....)))))).)))))..))))(((((((((.((((....)))))))))))))))))..--- ( -35.30, z-score = -4.21, R) >droVir3.scaffold_13049 2640849 84 - 25233164 -CCGUUGUACAAAGUGGAAAAUGUGGUUAAAAACC-GCAAUACGAAAUAAAUUACUUUUUCUGCUUUGAGCUGCAGUCCGAGGCUG--------------------- -..((.((.(((((..(((((((((((.....)))-))).................)))))..))))).)).))(((.....))).--------------------- ( -21.22, z-score = -1.99, R) >droMoj3.scaffold_6680 7551843 84 - 24764193 -CUGCUGUACAAAGUGGAAAAUGUGGUUAAAAACC-GCAAUACGAAAUAAAUUACUUUUUCUGCUUUGAGCUGCAGUCCGAGGCUG--------------------- -((((.((.(((((..(((((((((((.....)))-))).................)))))..))))).)).))))..........--------------------- ( -24.42, z-score = -3.17, R) >droGri2.scaffold_15110 20947860 85 + 24565398 -CUGUUGUACAAAGUGGAAAAUGUGGUUAAAAACC-GCAAUACGAAAUAAAUUACUUUUUCUGCUUUGCGCUGCAGUUCGAGGCUGU-------------------- -((((.(..(((((..(((((((((((.....)))-))).................)))))..)))))..).))))...........-------------------- ( -23.32, z-score = -2.36, R) >consensus _CUGUUGUACAAAGUGGAAAAUGUGGUUAAAAACC_GCAAUACGAAAUAAAUUACUUUUUCUGCUUUGAGCUGCAGUCCGAGACUCU_UGCCUUCUGCCU_______ .((((.((.(((((..(((((.((((((.....................)))))).)))))..))))).)).))))............................... (-20.96 = -20.97 + 0.01)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:22:19 2011