
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 12,806,922 – 12,807,023 |
| Length | 101 |
| Max. P | 0.776114 |

| Location | 12,806,922 – 12,807,023 |
|---|---|
| Length | 101 |
| Sequences | 13 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.18 |
| Shannon entropy | 0.43397 |
| G+C content | 0.57019 |
| Mean single sequence MFE | -37.84 |
| Consensus MFE | -20.39 |
| Energy contribution | -19.85 |
| Covariance contribution | -0.54 |
| Combinations/Pair | 1.37 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.776114 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr3L 12806922 101 - 24543557 -------------------AAAUAAAGUUUACAAAAUGGACUUCCAAAACCGCGCCGGCGGCAAGACCGGCAGCGGAGGCGUAGCCUCCUGGUCGGAGACGAACCGCGACCGCAAGGAGC -------------------.................(((....)))....(((((((((.....).)))))...((((((...)))))).(((((....)).)))))).((....))... ( -40.10, z-score = -2.44, R) >droPer1.super_20 285761 101 + 1872136 -------------------AAAAAAAACUUAAAAUAUGGACUUUCAAAAUCGUGCCGGUGGCAAAACCGGCAGCGGUGGCGUUGCCUCCUGGUCGGAGACCAACCGCGACCGCAAGGAGC -------------------......................((((.......(((((((......)))))))(((((.((((((.((((.....))))..))).))).)))))..)))). ( -36.80, z-score = -2.17, R) >dp4.chrXR_group6 1652866 100 + 13314419 --------------------AAAAAAACUUAAAAUAUGGACUUUCAAAAUCGUGCCGGUGGCAAAACCGGCAGCGGUGGCGUUGCCUCCUGGUCGGAGACCAACCGCGACCGCAAGGAGC --------------------.....................((((.......(((((((......)))))))(((((.((((((.((((.....))))..))).))).)))))..)))). ( -36.80, z-score = -2.13, R) >droAna3.scaffold_13337 3511505 101 + 23293914 -------------------AAAUAAUCUUGGAAAAAUGGACUUUCAGAACCGCGCCGGUGGCAAGACCGGCAGCGGAGGCGUUGCCUCCUGGUCGGAAUCGAACCGCGACCGCAAGGAAC -------------------......(((.((((........)))))))..(((((((((......))))))...((((((...)))))).(((((....)).)))))).((....))... ( -36.70, z-score = -1.39, R) >droEre2.scaffold_4784 12819218 101 - 25762168 -------------------AGAUAAAGCUCACGAAAUGGAUUUCCAAAACCGCGCCGGCGGCAAGACCGGCAGCGGAGGCGUGGCCUCCUGGUCGGAGACGAACCGCGACCGCAAGGAGC -------------------.......((.((((...(((....)))...((((((((((.....).))))).))))...))))))((((((((((....)).)))((....)).))))). ( -44.00, z-score = -2.43, R) >droSec1.super_0 4991550 101 - 21120651 -------------------AAAUAAAGUUUACAAAAUGGAUUUCCAAAACCGCGCCGGCGGCAAGACCGGCAGCGGAGGCGUGGCCUCCUGGUCGGAGACGAACCGCGACCGCAAGGAGC -------------------.................(((....)))....(((((((((.....).)))))...((((((...)))))).(((((....)).)))))).((....))... ( -40.10, z-score = -2.06, R) >droSim1.chr3L 12193670 101 - 22553184 -------------------AAAUAAAGUUUACAAAAUGGAUUUUCAGAACCGCGCCGGCGGCAAGACCGGCAGCGGAGGCGUGGCCUCCUGGUCGGAGACGAACCGCGACCGCAAGGAGC -------------------....((((((((.....))))))))......(((((((((.....).)))))...((((((...)))))).(((((....)).)))))).((....))... ( -38.50, z-score = -1.67, R) >droGri2.scaffold_15110 20819169 96 - 24565398 --------------------AAUUA---A-AAAACAUGGAUUUCCAAAAUCGCGCCGGUGGCAAAACCGGUAGCGGUGGUGUAGCCUCCUGGUCAGAGACCAAUCGCGAGCGCAAAGAAC --------------------.....---.-......(((....)))....(((((((((......)))))).((((((((.(.(((....)))...).))).)))))..)))........ ( -30.30, z-score = -1.74, R) >droMoj3.scaffold_6680 7433029 99 + 24764193 --------------------AGUUGAAUU-AAACCAUGGAUUUCCAAAAUCGCGCCGGUGGUAAAACCGGUAGCGGUGGCGUCGCCUCCUGGUCGGAGACAAACCGCGAGCGAAAGGAAC --------------------.........-...((.(((....)))...((((((((((......)))))).(((((...(((.((........)).)))..)))))..))))..))... ( -36.10, z-score = -1.91, R) >droVir3.scaffold_13049 2520096 99 + 25233164 --------------------AAUUAACUA-AAAACAUGGAUUUCCAGAAUCGCGCCGGUGGCAAAACCGGUAGCGGUGGCGUCGCCUCCUGGUCAGAGACAAAUCGCGAGCGAAAAGAAC --------------------.........-......(((....)))...((((((((((......)))))).(((((...((((((....)))....)))..)))))..))))....... ( -30.20, z-score = -1.28, R) >droWil1.scaffold_180698 5508320 100 - 11422946 --------------------AAUUAAAAACUCAACAUGGACUUCCAAAAUCGCGCCGGUGGCAAAACCGGUAGCGGUGGGGUGGCCUCCUGGUCGGAGACAAAUCGCGACCGCAAGGAAC --------------------........((((....(((....)))..(((((((((((......)))))).))))).))))..(((..((((((((......)).))))))..)))... ( -36.40, z-score = -2.06, R) >anoGam1.chr3L 14382346 120 - 41284009 AACUAACGUAAUUUCUCCCCACGCUUUCCAGUGCGAUGGAUUUCCAGAACCGAGCCGGUGGCAAAACCGGCGGCGGCGGUGUCGCCUCCUGGUCCGAAAGCAACCGGGAUCGGCGCGAGC .((....)).....(((.((((((........))).))).......(..((((((((((......))))))((((((...))))))(((((((.(....)..)))))))))))..)))). ( -48.00, z-score = -1.07, R) >triCas2.ChLG4 3333741 100 - 13894384 --------------------AAAUAAAAGUGCAAAAUGGAUUUCCAAAAUCGCCCGGGGGGCAAGACCGGCGGUGGCGGCGUAGCCUCCUGGUCGGAAACCAACCGCGACCGCCGCGAGC --------------------..........((....(((.(((((......((((...))))..((((((.((.(((......)))))))))))))))))))..((((.....)))).)) ( -37.90, z-score = -0.37, R) >consensus ____________________AAUAAAAUUUAAAAAAUGGAUUUCCAAAAUCGCGCCGGUGGCAAAACCGGCAGCGGUGGCGUGGCCUCCUGGUCGGAGACCAACCGCGACCGCAAGGAGC ....................................(((....)))...(((((((((........))))).))))..(((..(((....))).(....)....)))............. (-20.39 = -19.85 + -0.54)
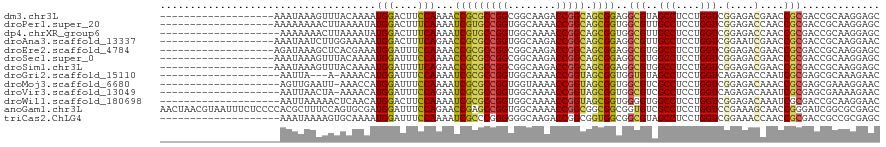
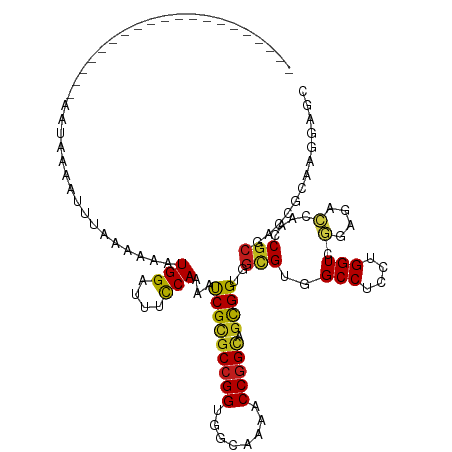
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:22:08 2011