
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 12,803,517 – 12,803,634 |
| Length | 117 |
| Max. P | 0.543480 |

| Location | 12,803,517 – 12,803,634 |
|---|---|
| Length | 117 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 72.48 |
| Shannon entropy | 0.58235 |
| G+C content | 0.60967 |
| Mean single sequence MFE | -45.27 |
| Consensus MFE | -15.69 |
| Energy contribution | -16.68 |
| Covariance contribution | 0.98 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.543480 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
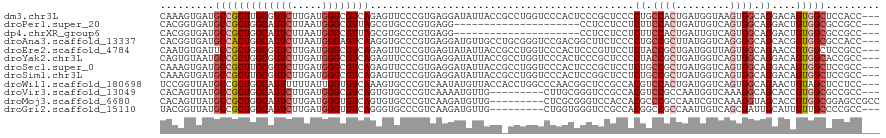
>dm3.chr3L 12803517 117 - 24543557 CAAAGUGAUGCCGCUUGCGUUCUUGAUGGGCGUCAGAGUUCCCGUGAGGAUAUUACCGCCUGGUCCCACUCCCGCUCCCCUGCCACUGAUGGUAAGUGGCAGGACAGUGGCUCCACC--- ....(((..((((((.(((........(((((....(((..((....))..)))..)))))((.......)))))...(((((((((.......)))))))))..))))))..))).--- ( -46.40, z-score = -2.27, R) >droPer1.super_20 282294 96 + 1872136 CACGGUGAUGCCGCUGGCAUUCUUAAUGGGCGUUAGCGUGCCCGUGAGG---------------------CCUCCUCCUCUUCCACUGAUUGUCAGUGGCAGGACUGUGGCGCCGCC--- ((((((..(((((((((((.((...(((((((......)))))))((((---------------------......)))).......)).)))))))))))..))))))........--- ( -45.00, z-score = -2.96, R) >dp4.chrXR_group6 1649397 96 + 13314419 CACGGUGAUGCCGCUGGCAUUCUUAAUGGGCGUUAGCGUGCCCGUGAGG---------------------CCUCCUCCUCUUCCACUGAUUGUCAGUGGCAGGACUGUGGCGCCGCC--- ((((((..(((((((((((.((...(((((((......)))))))((((---------------------......)))).......)).)))))))))))..))))))........--- ( -45.00, z-score = -2.96, R) >droAna3.scaffold_13337 3508014 117 + 23293914 CACGGUGAUGCCACUGGCAUUCUUAAUGGGAGUCAAGGUGCCCGUGAGGAUGUUGCCUGCGGGUCCGACGGCUUCUCCCCUGCCGCUUAUGGUCAGGGGCAGCACGGUGGCGCCACC--- ...((((.((((((((((.........(((((((..((.((((((.(((......)))))))))))...)))))))(((((((((....))).))))))..)).)))))))).))))--- ( -59.40, z-score = -2.89, R) >droEre2.scaffold_4784 12815770 117 - 25762168 CAAUGUGAUUCCGCUGGCGUUCUUGAUGGGCGUCAGAGUUCCCGUGAGUAUAUUACCGCCUGGUCCCACUCCCGUUCCUCUACCGCUGAUGGUUAGUGGCAGAACCGUGGCUCCGCC--- .(((((.(((((((((((((((.....)))))))))......)).)))))))))...(((((((.(((((.(((((...........)))))..)))))....)))).)))......--- ( -32.70, z-score = 0.72, R) >droYak2.chr3L 12878227 117 - 24197627 CAGUGUAAUGCCGCUGGCGUUCUUGAUGGGCGUCAGAGUUCCCGUGAGGAUAUUACCGCCUGGUCCCACUCCCGCUCCCCUACCGCUGAUGGUCAGUGGCAGGACAGUGGCACCGCC--- ..(((...(((((((((((........(((((....(((..((....))..)))..)))))((.......)))))...(((.((((((.....)))))).))).)))))))).))).--- ( -45.10, z-score = -0.84, R) >droSec1.super_0 4988152 117 - 21120651 CAAAGUGAUGCCGCUUGCGUUCUUGAUGGGCGUCAGAGUUCCCGUGAGGAUAUUACCGCCUGGUCCCACUCCCGCUCCUCUGCCGCUGAUGGUCAGUGGCAGGACAGUGGCUCCGCC--- ....(((..((((((...((((.....))))(((.((((....(((.((((...........)))))))....))))..(((((((((.....))))))))))))))))))..))).--- ( -46.00, z-score = -1.39, R) >droSim1.chr3L 12190183 117 - 22553184 CAAAGUGAUGCCGCUUGCGUUCUUGAUGGGCGUCAGAGUUCCCGUGAGGAUAUUACCGCCUGGUCCCACUCCGGCUCCUCUGCCGCUGAUGGUCAGUGGCAGGACAGUGGCUCCGCC--- ....(((..((((((...((((.....))))(((.(((((...(((.((((...........)))))))...)))))..(((((((((.....))))))))))))))))))..))).--- ( -46.80, z-score = -1.22, R) >droWil1.scaffold_180698 5910659 117 + 11422946 UCCGGUUAUGCCGCUGGCAUUUUUUAUUGGUGUCAAAGUGCCCGUCAAUAUGUUACCACCUGGCCCAACGGCUCCGCCACGUCCACUGAUGGUCAGUGGCAGAACUGUAGCUCCUCC--- ..(((((.((((((((((........((((.((((..(((..(((....)))....))).)))))))).(((...))).((((....)))))))))))))).)))))..........--- ( -36.20, z-score = -0.97, R) >droVir3.scaffold_13049 2516109 108 + 25233164 CACAGUUAUGCCGCUGGCAUUCUUGAUGGGCGUCAGUGUGCCCGUCAAAAUGUUG---------CUUGCGGGUCCGCCACGUCCGCCAAUGGUCAAAGGCAGCACCGUGGCGCCGCC--- ..........((((.((((...((((((((((......)))))))))).....))---------)).))))...(((((((.(.(((..........))).)...))))))).....--- ( -42.80, z-score = -0.58, R) >droMoj3.scaffold_6680 7428700 111 + 24764193 CACAGUUAUGCCGCUGGCAUUCUUGAUGGGUGUCAGUGUGCCCGUCAAGAUGUUG---------CUCGCGGGUCCACCACGCCCGCCAAUCGUCAAAGGUAGCACCGUGGCGGAGCCGCC ..((((......))))(((.((((((((((..(....)..))))))))))...))---------)..((((.(((.(((((...((..(((......))).))..))))).))).)))). ( -49.60, z-score = -2.69, R) >droGri2.scaffold_15110 20815329 108 - 24565398 UACGGUUAUGCCGCUGGCAUUCUUGAUGGGUGUCAGGGUGCCCGUCAAGAUGUUG---------CUGGUGGGUCCGCCACGGCCGCCAAUUGUCAGCGAUUGCAUUGUUGCCCCGCC--- ...(((.((((((((((((.((((((((((..(....)..)))))))))).((((---------.(((((....))))))))).......))))))))...))))....))).....--- ( -48.20, z-score = -2.17, R) >consensus CACAGUGAUGCCGCUGGCAUUCUUGAUGGGCGUCAGAGUGCCCGUGAGGAUAUUACC_CCUGGUCCCACGCCUCCUCCUCUGCCGCUGAUGGUCAGUGGCAGGACAGUGGCGCCGCC___ .........(((((((((((((.....))))))))............................................((.((((.........)))).))....)))))......... (-15.69 = -16.68 + 0.98)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:22:07 2011