| Sequence ID | dm3.chr3L |
|---|---|
| Location | 12,791,064 – 12,791,154 |
| Length | 90 |
| Max. P | 0.998172 |

| Location | 12,791,064 – 12,791,154 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 72.82 |
| Shannon entropy | 0.51800 |
| G+C content | 0.33399 |
| Mean single sequence MFE | -22.49 |
| Consensus MFE | -9.71 |
| Energy contribution | -10.10 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.47 |
| Mean z-score | -3.32 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.28 |
| SVM RNA-class probability | 0.998172 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
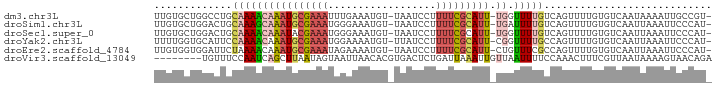
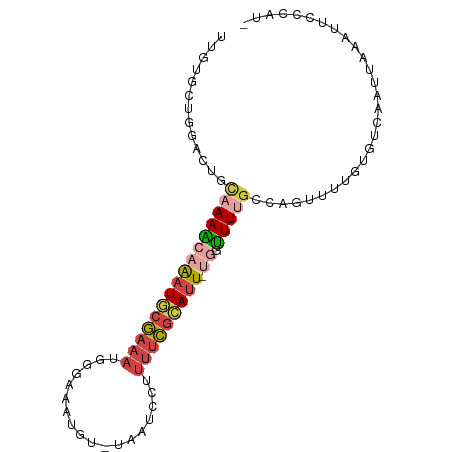
>dm3.chr3L 12791064 90 + 24543557 -ACGGCAAUUUUAUUGACACAAAACUGACAAAACCA-AAUGCGAAAAGGAUUA-ACAUUUCAAAUUUCGCAUUUGUUUUGCAGGCCAGCACAA -..(((.......(((...)))..(((.((((((.(-(((((((((..((...-.....))...))))))))))))))))))))))....... ( -25.60, z-score = -4.54, R) >droSim1.chr3L 12178195 90 + 22553184 -AUGGGAAUUUAAUUGACACAAAACUGACAAAAUCA-AAUGCGAAAAGGAUUA-ACAUUUCCCAUUUCGCAUUUGCUUUGCAGUCCAGCACAA -.(((........(((...))).((((.((((..((-(((((((((.(((...-.....)))..))))))))))).)))))))))))...... ( -22.90, z-score = -3.18, R) >droSec1.super_0 4975954 90 + 21120651 -AUGGGAAUUUAAUUGACACAAAACUGACAAAACCA-AAUGCGAAAAGGAUUA-ACAUUUCCCAUUUCGUAUUUGUUUUGCAGUCCAGCACAA -.(((........(((...))).((((.((((((.(-(((((((((.(((...-.....)))..)))))))))))))))))))))))...... ( -23.20, z-score = -3.29, R) >droYak2.chr3L 12862554 90 + 24197627 -AUGGGAAUUUAAUUGACACAAAACUGGCAAAACCG-AAUGCGAAAAGGAUAA-ACAUUUUCCAUUUCGCAUUUGUUUUGGAAUGCACCAAAA -.(((..((((........((....)).((((((.(-(((((((((.(((...-......))).))))))))))))))))))))...)))... ( -24.10, z-score = -3.16, R) >droEre2.scaffold_4784 12798694 90 + 25762168 -AUGGGAAUUUAAUUGACACAAAACUGGCGAAACAG-AAUGCGAAAAGGAUUA-ACAUUUUCUAUUUCGCAUUUGUUUUAGAAUCCACCACAA -...(((.(((((..((((.....(((......)))-(((((((((((((...-.....)))).)))))))))))))))))).)))....... ( -21.60, z-score = -3.48, R) >droVir3.scaffold_13049 2505488 85 - 25233164 UCUGUUACUUUUAUUAACGAAAGUUUGGAAAAUUAACAAUUUAAUCAGAGUCACGUGUUAAUUACUAUUAAGCUGAUUGGAAACA-------- ..(((((.((((...(((....)))...)))).))))).(((((((((......(((.....))).......)))))))))....-------- ( -17.52, z-score = -2.27, R) >consensus _AUGGGAAUUUAAUUGACACAAAACUGACAAAACCA_AAUGCGAAAAGGAUUA_ACAUUUCCCAUUUCGCAUUUGUUUUGCAAUCCAGCACAA ............................((((((...(((((((((.((............)).))))))))).))))))............. ( -9.71 = -10.10 + 0.39)


| Location | 12,791,064 – 12,791,154 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 72.82 |
| Shannon entropy | 0.51800 |
| G+C content | 0.33399 |
| Mean single sequence MFE | -22.35 |
| Consensus MFE | -7.57 |
| Energy contribution | -8.27 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.40 |
| Mean z-score | -2.82 |
| Structure conservation index | 0.34 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.64 |
| SVM RNA-class probability | 0.957633 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 12791064 90 - 24543557 UUGUGCUGGCCUGCAAAACAAAUGCGAAAUUUGAAAUGU-UAAUCCUUUUCGCAUU-UGGUUUUGUCAGUUUUGUGUCAAUAAAAUUGCCGU- .......((((((((((((((((((((((...((.....-...))..)))))))))-).)))))).)))((((((....))))))..)))..- ( -25.60, z-score = -3.35, R) >droSim1.chr3L 12178195 90 - 22553184 UUGUGCUGGACUGCAAAGCAAAUGCGAAAUGGGAAAUGU-UAAUCCUUUUCGCAUU-UGAUUUUGUCAGUUUUGUGUCAAUUAAAUUCCCAU- ((((((..(((((((((((((((((((((.((((.....-...)))))))))))))-)).))))).)))))..))).)))............- ( -30.70, z-score = -4.72, R) >droSec1.super_0 4975954 90 - 21120651 UUGUGCUGGACUGCAAAACAAAUACGAAAUGGGAAAUGU-UAAUCCUUUUCGCAUU-UGGUUUUGUCAGUUUUGUGUCAAUUAAAUUCCCAU- ((((((..(((((((((((((((.(((((.((((.....-...))))))))).)))-).)))))).)))))..))).)))............- ( -24.10, z-score = -2.75, R) >droYak2.chr3L 12862554 90 - 24197627 UUUUGGUGCAUUCCAAAACAAAUGCGAAAUGGAAAAUGU-UUAUCCUUUUCGCAUU-CGGUUUUGCCAGUUUUGUGUCAAUUAAAUUCCCAU- ..((((..((...((((((.(((((((((.(((......-...))).)))))))))-..)))))).......))..))))............- ( -23.10, z-score = -2.79, R) >droEre2.scaffold_4784 12798694 90 - 25762168 UUGUGGUGGAUUCUAAAACAAAUGCGAAAUAGAAAAUGU-UAAUCCUUUUCGCAUU-CUGUUUCGCCAGUUUUGUGUCAAUUAAAUUCCCAU- ..((((.(((((.....(((((.((((((((((..((((-.((.....)).)))))-)))))))))....)))))........)))))))))- ( -22.52, z-score = -3.71, R) >droVir3.scaffold_13049 2505488 85 + 25233164 --------UGUUUCCAAUCAGCUUAAUAGUAAUUAACACGUGACUCUGAUUAAAUUGUUAAUUUUCCAAACUUUCGUUAAUAAAAGUAACAGA --------.......((((((.(((...((.....))...)))..))))))...((((((.((((...(((....)))...)))).)))))). ( -8.10, z-score = 0.40, R) >consensus UUGUGCUGGACUGCAAAACAAAUGCGAAAUGGGAAAUGU_UAAUCCUUUUCGCAUU_UGGUUUUGCCAGUUUUGUGUCAAUUAAAUUCCCAU_ .............((((((.(((((((((..................)))))))))...))))))............................ ( -7.57 = -8.27 + 0.70)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:22:05 2011