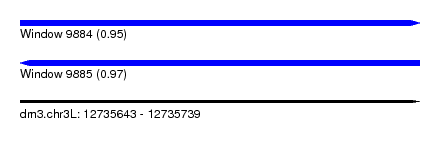
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 12,735,643 – 12,735,739 |
| Length | 96 |
| Max. P | 0.973693 |

| Location | 12,735,643 – 12,735,739 |
|---|---|
| Length | 96 |
| Sequences | 7 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 60.54 |
| Shannon entropy | 0.77810 |
| G+C content | 0.37404 |
| Mean single sequence MFE | -17.62 |
| Consensus MFE | -6.78 |
| Energy contribution | -8.26 |
| Covariance contribution | 1.47 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.949821 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
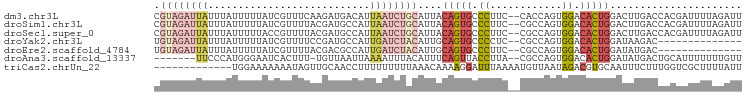
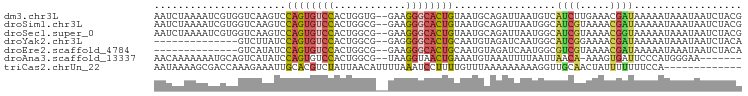
>dm3.chr3L 12735643 96 + 24543557 CGUAGAUUAUUUAUUUUUAUCGUUUCAAGAUGACAUUAAUCUGCAUUACAGUGCCCUUC--CACCAGUGGACACUGGACUUGACCACGAUUUUAGAUU .((((((((.......(((((.......)))))...))))))))....(((((....((--((....)))))))))...................... ( -20.10, z-score = -2.22, R) >droSim1.chr3L 12119600 96 + 22553184 CGUAGAUUAUUUAUUUUUAUCGUUUUACGAUGCCAUUAAUCUGCAUUACAGUGCCCUUC--CGCCAGUGGACACUGGACUUGACCACGAUUUUAGAUU .((((((((........(((((.....)))))....))))))))....(((((((((..--....)).)).)))))...................... ( -20.60, z-score = -2.30, R) >droSec1.super_0 4920684 96 + 21120651 CGUAGAUUAUUUAUUUUUACCGUUUUACGAUGCCAUUAAUCUGCAUUACAGUGCCCUUC--CGCCAGUGGACACUGGACUUGACCACGAUUUUAGAUU .((((((((...........((.....)).......))))))))....(((((((((..--....)).)).)))))...................... ( -16.37, z-score = -0.97, R) >droYak2.chr3L 12807035 82 + 24197627 UGUAGAUUAUUUAUUUUUAUCGUUUUCCGAUGCCAUUGAUCUACAUUGCAGUGCCCCUC--CGCCAGUGGACACUGGAUAAGAC-------------- (((((((((........(((((.....)))))....)))))))))...(((((((.((.--....)).)).)))))........-------------- ( -19.50, z-score = -2.11, R) >droEre2.scaffold_4784 12743305 82 + 25762168 UGUAGAUUAUUUAUUUUUAUCGUUUUACGACGCCAUUGAUCUACAUUGCAGUGCCCUUC--CGCCAGUGGACACUGGAUAUGAC-------------- (((((((((..........(((.....)))......)))))))))...(((((((((..--....)).)).)))))........-------------- ( -18.19, z-score = -1.74, R) >droAna3.scaffold_13337 20006626 88 - 23293914 -------UUCCCAUGGGAAUCACUUU-UGUUAAUUAAAAUUUACAUUUCAGUUACCUUA--CGCCAGUGGACACUGGAUAUGACUGCAUUUUUUUGUU -------((((....)))).......-.....................((((((.....--..(((((....)))))...))))))............ ( -14.40, z-score = -0.22, R) >triCas2.chrUn_22 323957 85 + 327011 -------------UGGAAAAAAAUAGUUGCAACCUUUUUUUUUAAACAAAAGGAUUUAAAAUGUUAAUAGACGUGCAAUUUCUUUGGUCGCUUUUAUU -------------..((..(((..((((((..((((((.........)))))).......(((((....)))))))))))..)))..))......... ( -14.20, z-score = -0.71, R) >consensus CGUAGAUUAUUUAUUUUUAUCGUUUUACGAUGCCAUUAAUCUACAUUACAGUGCCCUUC__CGCCAGUGGACACUGGAUUUGACCACGAUUUUAGAUU .((((((((...........................))))))))....(((((.((............)).)))))...................... ( -6.78 = -8.26 + 1.47)
| Location | 12,735,643 – 12,735,739 |
|---|---|
| Length | 96 |
| Sequences | 7 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 60.54 |
| Shannon entropy | 0.77810 |
| G+C content | 0.37404 |
| Mean single sequence MFE | -18.09 |
| Consensus MFE | -8.07 |
| Energy contribution | -8.06 |
| Covariance contribution | -0.02 |
| Combinations/Pair | 1.58 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.89 |
| SVM RNA-class probability | 0.973693 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
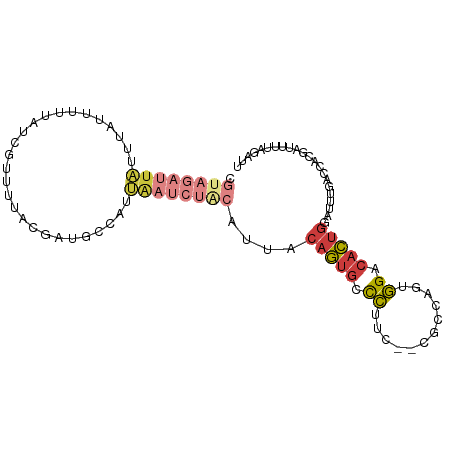
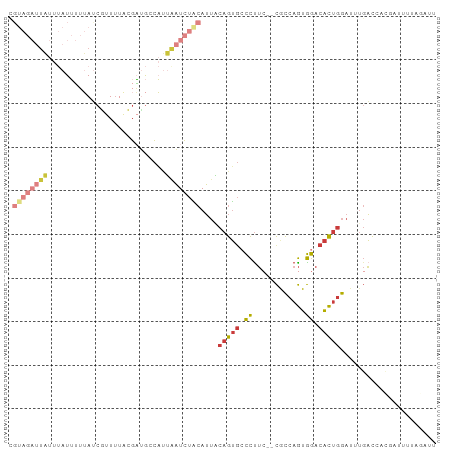
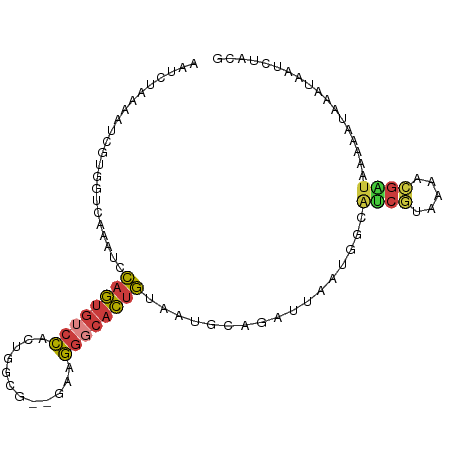
>dm3.chr3L 12735643 96 - 24543557 AAUCUAAAAUCGUGGUCAAGUCCAGUGUCCACUGGUG--GAAGGGCACUGUAAUGCAGAUUAAUGUCAUCUUGAAACGAUAAAAAUAAAUAAUCUACG ........(((((..(((((..((((((((.......--...)))))))).......(((....)))..))))).))))).................. ( -22.90, z-score = -2.50, R) >droSim1.chr3L 12119600 96 - 22553184 AAUCUAAAAUCGUGGUCAAGUCCAGUGUCCACUGGCG--GAAGGGCACUGUAAUGCAGAUUAAUGGCAUCGUAAAACGAUAAAAAUAAAUAAUCUACG ..........(((((....(((((((((((.......--...))))))))((((....))))..)))((((.....)))).............))))) ( -19.30, z-score = -1.04, R) >droSec1.super_0 4920684 96 - 21120651 AAUCUAAAAUCGUGGUCAAGUCCAGUGUCCACUGGCG--GAAGGGCACUGUAAUGCAGAUUAAUGGCAUCGUAAAACGGUAAAAAUAAAUAAUCUACG ..........(((((....(((((((((((.......--...))))))))((((....))))..)))((((.....)))).............))))) ( -18.70, z-score = -0.64, R) >droYak2.chr3L 12807035 82 - 24197627 --------------GUCUUAUCCAGUGUCCACUGGCG--GAGGGGCACUGCAAUGUAGAUCAAUGGCAUCGGAAAACGAUAAAAAUAAAUAAUCUACA --------------........((((((((.((....--.))))))))))...(((((((.......((((.....))))...........))))))) ( -20.37, z-score = -1.78, R) >droEre2.scaffold_4784 12743305 82 - 25762168 --------------GUCAUAUCCAGUGUCCACUGGCG--GAAGGGCACUGCAAUGUAGAUCAAUGGCGUCGUAAAACGAUAAAAAUAAAUAAUCUACA --------------........((((((((.......--...))))))))...(((((((.......((((.....))))...........))))))) ( -18.47, z-score = -1.40, R) >droAna3.scaffold_13337 20006626 88 + 23293914 AACAAAAAAAUGCAGUCAUAUCCAGUGUCCACUGGCG--UAAGGUAACUGAAAUGUAAAUUUUAAUUAACA-AAAGUGAUUCCCAUGGGAA------- ..............(((((..(((((....)))))..--...(.(((.((((((....)))))).))).).-...)))))(((....))).------- ( -14.70, z-score = -0.24, R) >triCas2.chrUn_22 323957 85 - 327011 AAUAAAAGCGACCAAAGAAAUUGCACGUCUAUUAACAUUUUAAAUCCUUUUGUUUAAAAAAAAAGGUUGCAACUAUUUUUUUCCA------------- .......((((((.............((......)).((((((((......)))))))).....))))))...............------------- ( -12.20, z-score = -1.20, R) >consensus AAUCUAAAAUCGUGGUCAAAUCCAGUGUCCACUGGCG__GAAGGGCACUGUAAUGCAGAUUAAUGGCAUCGUAAAACGAUAAAAAUAAAUAAUCUACG ......................((((((((............)))))))).................((((.....)))).................. ( -8.07 = -8.06 + -0.02)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:21:59 2011