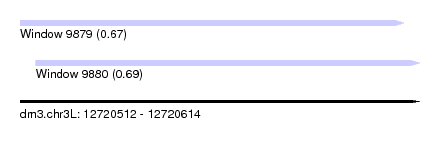
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 12,720,512 – 12,720,614 |
| Length | 102 |
| Max. P | 0.691785 |

| Location | 12,720,512 – 12,720,610 |
|---|---|
| Length | 98 |
| Sequences | 12 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 72.55 |
| Shannon entropy | 0.54937 |
| G+C content | 0.48838 |
| Mean single sequence MFE | -22.87 |
| Consensus MFE | -10.70 |
| Energy contribution | -10.51 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.671548 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 12720512 98 + 24543557 --------------CGGGACGAACACCCCCC-GAGGAUGCCCUCGCUG---CAGUGCUGCAAAUGAUGAGUUGUGCCUAAUUCAUAACCACAUCGAACG-AAUUAACUCGCAGUAUG --------------.(((........))).(-((((....)))))...---..(((((((...(((((.((((((.......))))))..)))))...(-(......))))))))). ( -28.00, z-score = -2.32, R) >droSim1.chr3L 12104775 98 + 22553184 --------------CGGGACGACCACCCCCC-GAGGAUGCCCUCGAUG---CAGUGCUGCAAAUGAUGAGUUGUGCCUAAUUCAUAACCACAUCGAACG-AAUUAACUCGCAGUAUG --------------.(((........))).(-((((....)))))...---..(((((((...(((((.((((((.......))))))..)))))...(-(......))))))))). ( -28.70, z-score = -2.72, R) >droSec1.super_0 4905895 98 + 21120651 --------------CGGGACGACCACCCCCC-GACGAUGCCCUCGAUG---CAGUGCUGCAAAUGAUGAGUUGUGCCUAAUUCAUAACCACAUCGAACG-AAUUAACUCGCAGUAUG --------------((((..........)))-).(((.....)))...---..(((((((...(((((.((((((.......))))))..)))))...(-(......))))))))). ( -23.80, z-score = -1.57, R) >droYak2.chr3L 12791478 94 + 24197627 ----------------------CCACCCCGCAAAGGAAGCGUUCGAUGCUGCAGUGCUGCAAAUGAUGAGUUGUGCCUAAUUCAUAACCACAUCGAACG-AAUUAACUCGCAGCAUG ----------------------.......((....((..(((((((((.((((....))))....((((((((....)))))))).....)))))))))-..)).....))...... ( -24.10, z-score = -1.28, R) >droEre2.scaffold_4784 12727001 87 + 25762168 ----------------------CCACCCCC--AAGGA--UGUUCGAUG---CAGUGCUGCAGAUGAUGAGUUGUGCCUAAUUCAUAACCACAUCGAACG-AAUUAACUCGCAGCAUG ----------------------...((...--..)).--(((.....)---))(((((((.((((.((.((((((.......))))))))))))....(-(......))))))))). ( -22.30, z-score = -1.84, R) >droAna3.scaffold_13337 19988144 94 - 23293914 ---------------AGGAAAGCCCACCCCC--GUCGAACAGACGUCG-----AUGCUGCAAAUGAUGAGUUGUGCCUAAUUCAUAACCACAUCGAACG-AAUUAACUCACAGUAUG ---------------.((.....))..(..(--(((.....))))..)-----((((((....(((((.((((((.......))))))..)))))...(-(......)).)))))). ( -18.70, z-score = -1.15, R) >dp4.chrXR_group6 3512207 94 + 13314419 -----------------AAGAGCUGGCGACUGGGGCCCGCCCUCGCUG-----AUGCUGCAAAUGAUGAGUUGUGCCUAAUUCAUAACCACAUCGAACG-AAUUAACUCGCAGUAUG -----------------.......(((((...(((....)))))))).-----(((((((...(((((.((((((.......))))))..)))))...(-(......))))))))). ( -27.80, z-score = -1.63, R) >droPer1.super_9 1807814 94 + 3637205 -----------------AAGAGUUGGCGACUGGGGCCCGCCCUCACUG-----AUGCUGCAAAUGAUGAGUUGUGCCUAAUUCAUAACCACAUCGAACG-AAUUAACUCGCAGUAUG -----------------..(((..((((.........)))))))....-----(((((((...(((((.((((((.......))))))..)))))...(-(......))))))))). ( -25.20, z-score = -1.24, R) >droWil1.scaffold_181096 11879226 94 - 12416693 ----------------CGGACAUCCCACCCUUUAUUAAGCAAACACUA-----AAGCUGCAAAUGAUGCUUUGUGCCGAAUUCAUAACCAUAUCGAAUG-GAUUUACU-GCAGGAUA ----------------.((.....))......((((..(((..(((.(-----((((..........))))))))..((((((((...........)))-)))))..)-))..)))) ( -14.90, z-score = 0.15, R) >droVir3.scaffold_13049 19091562 101 + 25233164 ---------CGGUCCGCCCCC-CCACCCCAACAACGC-ACAGUCGCAG-----AUGCUGCAAAUGAUGAGUUGUGCCUAAUUCAUAACCACAUCGAAAGGAAUUAACUCGCAGCAUG ---------.((....))...-.............((-......))..-----(((((((...(((((.((((((.......))))))..)))))....((......))))))))). ( -21.60, z-score = -1.00, R) >droMoj3.scaffold_6680 6836250 99 + 24764193 ---------CGGUCCG--CCCACCCACCUAA-AAAGC-ACAGUCGCAG-----AUGCUGCAAAUGAUGAGUUGUGCCUAAUUCAUAACCACAUCGAAAGGAAUUAACUCACAGCAUG ---------.(((...--...))).......-...((-......))..-----((((((.......(((((((..(((..(((...........))))))...))))))))))))). ( -18.31, z-score = -0.26, R) >droGri2.scaffold_15110 16776689 112 + 24565398 CAAUCCGCCCAUCCUAUUCCCACCAAUCCACCAAUGCUGCAGUCACAG-----AUGCUGCAAAUGAUGAGUUGUGCCUAAUUCAUAACCACAUCGAAAGGAAUUAACUCACAGCAUG .................................((((((((((.....-----..))))))..((.(((((((..(((..(((...........))))))...))))))))))))). ( -21.00, z-score = -1.27, R) >consensus _________________GACCACCACCCCCC_AAGGAUGCACUCGCUG_____AUGCUGCAAAUGAUGAGUUGUGCCUAAUUCAUAACCACAUCGAACG_AAUUAACUCGCAGUAUG .....................................................((((((.....((((.((((((.......))))))..))))((...........)).)))))). (-10.70 = -10.51 + -0.19)
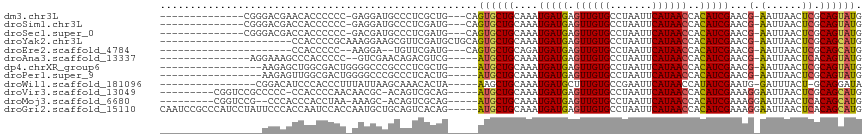
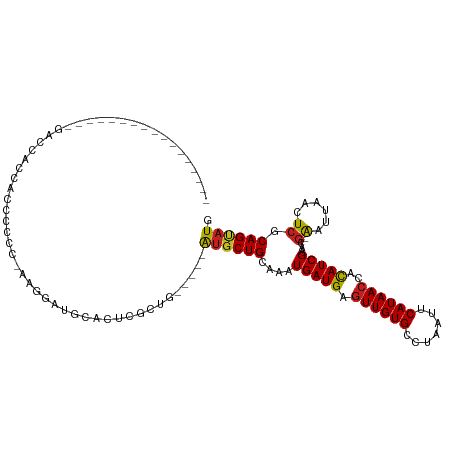
| Location | 12,720,516 – 12,720,614 |
|---|---|
| Length | 98 |
| Sequences | 12 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 74.43 |
| Shannon entropy | 0.50656 |
| G+C content | 0.48416 |
| Mean single sequence MFE | -23.36 |
| Consensus MFE | -10.93 |
| Energy contribution | -10.73 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.691785 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 12720516 98 + 24543557 --------------ACGAACACCCC-CCGAGGAUGCCCUC-GCUGC---AGUGCUGCAAAUGAUGAGUUGUGCCUAAUUCAUAACCACAUCGAACG-AAUUAACUCGCAGUAUGGAAC --------------.......((..-.(((((....))))-)....---.(((((((...(((((.((((((.......))))))..)))))...(-(......)))))))))))... ( -25.50, z-score = -2.23, R) >droSim1.chr3L 12104779 98 + 22553184 --------------ACGACCACCCC-CCGAGGAUGCCCUC-GAUGC---AGUGCUGCAAAUGAUGAGUUGUGCCUAAUUCAUAACCACAUCGAACG-AAUUAACUCGCAGUAUGGAAC --------------....((.....-.(((((....))))-)....---.(((((((...(((((.((((((.......))))))..)))))...(-(......)))))))))))... ( -27.10, z-score = -2.95, R) >droSec1.super_0 4905899 98 + 21120651 --------------ACGACCACCCC-CCGACGAUGCCCUC-GAUGC---AGUGCUGCAAAUGAUGAGUUGUGCCUAAUUCAUAACCACAUCGAACG-AAUUAACUCGCAGUAUGGAAC --------------....((.....-....(((.....))-)....---.(((((((...(((((.((((((.......))))))..)))))...(-(......)))))))))))... ( -20.70, z-score = -1.37, R) >droYak2.chr3L 12791478 98 + 24197627 ------------------CCACCCCGCAAAGGAAGCGUUC-GAUGCUGCAGUGCUGCAAAUGAUGAGUUGUGCCUAAUUCAUAACCACAUCGAACG-AAUUAACUCGCAGCAUGGAAC ------------------.....((.....))....((((-.((((((((((........(((((.((((((.......))))))..)))))....-.....))).))))))).)))) ( -28.13, z-score = -2.03, R) >droEre2.scaffold_4784 12727001 91 + 25762168 ------------------CCACCCC--CAAGGA--UGUUC-GAUGC---AGUGCUGCAGAUGAUGAGUUGUGCCUAAUUCAUAACCACAUCGAACG-AAUUAACUCGCAGCAUGGAAC ------------------......(--(.....--(((..-...))---)(((((((.((((.((.((((((.......))))))))))))....(-(......)))))))))))... ( -23.80, z-score = -1.75, R) >droAna3.scaffold_13337 19988148 94 - 23293914 -------------------AAGCCCACCCCCGUCGAACA--GACGU--CGAUGCUGCAAAUGAUGAGUUGUGCCUAAUUCAUAACCACAUCGAACG-AAUUAACUCACAGUAUGGAAC -------------------...........((((.....--))))(--(.((((((....(((((.((((((.......))))))..)))))...(-(......)).)))))).)).. ( -20.70, z-score = -1.78, R) >dp4.chrXR_group6 3512211 94 + 13314419 ---------------------GCUGGCGACUGGGGCCCGCCCUCGC--UGAUGCUGCAAAUGAUGAGUUGUGCCUAAUUCAUAACCACAUCGAACG-AAUUAACUCGCAGUAUGGAAC ---------------------...(((((...(((....)))))))--).(((((((...(((((.((((((.......))))))..)))))...(-(......)))))))))..... ( -27.80, z-score = -1.57, R) >droPer1.super_9 1807818 94 + 3637205 ---------------------GUUGGCGACUGGGGCCCGCCCUCAC--UGAUGCUGCAAAUGAUGAGUUGUGCCUAAUUCAUAACCACAUCGAACG-AAUUAACUCGCAGUAUGGAAC ---------------------(..((((.........))))..).(--(.(((((((...(((((.((((((.......))))))..)))))...(-(......)))))))))))... ( -23.40, z-score = -0.46, R) >droWil1.scaffold_181096 11879230 94 - 12416693 -------------------CAUCCCACC-CUUUAUUAAGCAAACAC--UAAAGCUGCAAAUGAUGCUUUGUGCCGAAUUCAUAACCAUAUCGAAUG-GAUUUACU-GCAGGAUAGAAC -------------------.........-.((((((..(((..(((--.(((((..........))))))))..((((((((...........)))-)))))..)-))..)))))).. ( -16.30, z-score = -0.90, R) >droVir3.scaffold_13049 19091566 101 + 25233164 ---------------CCGCCCCCCCACCCCAACAACGCACAGUCGC--AGAUGCUGCAAAUGAUGAGUUGUGCCUAAUUCAUAACCACAUCGAAAGGAAUUAACUCGCAGCAUGGAGC ---------------..((.((..............((......))--..(((((((...(((((.((((((.......))))))..)))))....((......))))))))))).)) ( -23.50, z-score = -1.51, R) >droMoj3.scaffold_6680 6836254 99 + 24764193 ---------------CCGCCCACCCACCUAAAAAGCACA--GUCGC--AGAUGCUGCAAAUGAUGAGUUGUGCCUAAUUCAUAACCACAUCGAAAGGAAUUAACUCACAGCAUGUAGC ---------------...................((...--...))--....((((((..((.(((((((..(((..(((...........))))))...)))))))))...)))))) ( -19.70, z-score = -0.99, R) >droGri2.scaffold_15110 16776693 112 + 24565398 CCGCCCAUCCUAUUCCCACCAAUCCACC--AAUGCUGCA--GUCAC--AGAUGCUGCAAAUGAUGAGUUGUGCCUAAUUCAUAACCACAUCGAAAGGAAUUAACUCACAGCAUGGAUC ..........................((--(.(((((((--((...--....))))))..((.(((((((..(((..(((...........))))))...)))))))))))))))... ( -23.70, z-score = -1.30, R) >consensus __________________CCACCCCACCAAGGAAGCCCUC_GACGC___AAUGCUGCAAAUGAUGAGUUGUGCCUAAUUCAUAACCACAUCGAACG_AAUUAACUCGCAGUAUGGAAC ..................................................((((((.....((((.((((((.......))))))..))))((...........)).))))))..... (-10.93 = -10.73 + -0.19)

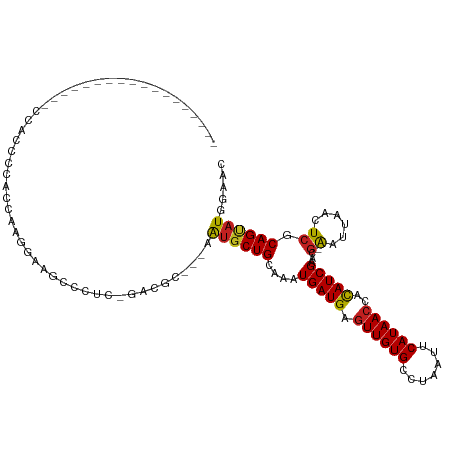
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:21:55 2011