
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 12,715,572 – 12,715,675 |
| Length | 103 |
| Max. P | 0.842061 |

| Location | 12,715,572 – 12,715,675 |
|---|---|
| Length | 103 |
| Sequences | 11 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 64.17 |
| Shannon entropy | 0.74438 |
| G+C content | 0.47657 |
| Mean single sequence MFE | -32.50 |
| Consensus MFE | -8.30 |
| Energy contribution | -7.74 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.26 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.835309 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
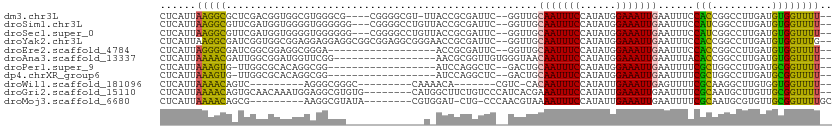
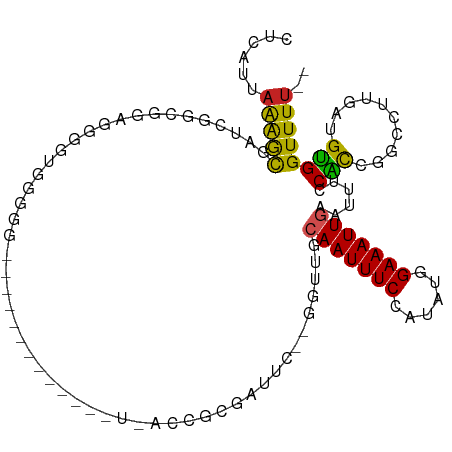
>dm3.chr3L 12715572 103 + 24543557 CUCAUUAAGGCGCUCGACGGUGGCGUGGGCG----CGGGGCGU-UUACCGCGAUUC--GGUUGCAAUUUCCAUAUGGAAAUUGAAUUUCCACCGGCCUUGAUGUGGUUUU-- (.(((((((((......((((((((....((----(((.....-...)))))...)--)....((((((((....)))))))).....))))))))))))))).).....-- ( -42.00, z-score = -2.63, R) >droSim1.chr3L 12099858 105 + 22553184 CUCAUUAAGGCGUUCGAUGGUGGGGUGGGGGG---CGGGGCCUGUUACCGCGAUUC--GGUUGCAAUUUCCAUAUGGAAAUUGAAUUUCCAUCGGCCUUGAUGUGGUUUU-- (.(((((((((...((((((((.(((((.(((---(...)))).))))).))....--.....((((((((....)))))))).....))))))))))))))).).....-- ( -44.20, z-score = -3.71, R) >droSec1.super_0 4900995 105 + 21120651 CUCAUUAAGGCGUUCGAUGGUGGGGUGGGGGG---CGGGGCCUGUUACCGCGAUUC--GGUUGCAAUUUCCAUAUGGAAAUUGAAUUUCCAUCGGCCUUGAUGUGGUUUU-- (.(((((((((...((((((((.(((((.(((---(...)))).))))).))....--.....((((((((....)))))))).....))))))))))))))).).....-- ( -44.20, z-score = -3.71, R) >droYak2.chr3L 12786490 108 + 24197627 CUCAUUAAGGCGAUCGGUGGCGGAGGAGGAGGCGGCGGAGGCGGGAACCGCGAUUC--GGUUGCAAUUUCCAUAUGGAAAUUGAAUUUCCACCGGCCUUGAUGUGGUUUG-- (.(((((((((...((((((..(........(((((.(((((((...))))..)))--.)))))(((((((....)))))))...)..))))))))))))))).).....-- ( -43.00, z-score = -2.59, R) >droEre2.scaffold_4784 12722291 90 + 25762168 CUCAUUAGGGCGAUCGGCGGAGGCGGGA------------------ACCGCGAUUC--GGUUGCAAUUUCCAUAUGGAAAUUGAAUUUCCACCGGCCUUGAUGUGGUUUU-- (.(((((((((...(((.((((((((..------------------.)))).....--.....((((((((....))))))))...)))).)))))))))))).).....-- ( -35.80, z-score = -2.74, R) >droAna3.scaffold_13337 19983377 93 - 23293914 CUCAUUAAAACGAUUGGCGGAUGGUUCGG-----------------AACGCGGUUGUGGGUAACAAUUUCCAUAUGGAAAUUGAAUUUACACCGGCCUUGAUGUGGUUUU-- ......(((((.((.(((.((....))..-----------------....((((.((((((..((((((((....)))))))).))))))))))))).....)).)))))-- ( -25.00, z-score = -1.04, R) >droPer1.super_9 1803654 89 + 3637205 CUCAUUAAAGUG-UUGGCGCACAGGCGG------------------AUCCAGGCUC--GACUGCAAUUUCCAUAUGGAAAUUGAAUUUUCGCUGGCCUUGAUGCGGUUUU-- .........(((-(....))))...((.------------------(((.((((((--((...((((((((....)))))))).....)))..))))).))).)).....-- ( -27.80, z-score = -1.44, R) >dp4.chrXR_group6 3508081 89 + 13314419 CUCAUUAAAGUG-UUGGCGCACAGGCGG------------------AUCCAGGCUC--GACUGCAAUUUCCAUAUGGAAAUUGAAUUUUCGCUGGCCUUGAUGCGGUUUU-- .........(((-(....))))...((.------------------(((.((((((--((...((((((((....)))))))).....)))..))))).))).)).....-- ( -27.80, z-score = -1.44, R) >droWil1.scaffold_181096 11674647 84 - 12416693 CUCAUUAAAACAGUC---------AGGGCGGGC---------CAAAACA-------CGUC-CACAAUUUCCAUAUUGAAAUUGAGUUUUCGCAAGGCUUGUGGUGGUUUU-- .((((((....((((---------.((((((..---------.....).-------))))-).(((((((......)))))))...........))))..))))))....-- ( -19.70, z-score = -0.78, R) >droGri2.scaffold_15110 7785130 102 - 24565398 CUCAUUAAAACAGUGCAACAAAUGGAGGCGUGUG--------CAUGGCUUCUGUCCCAUCACGAAAUUUCCAUAUUGAAAUUGAAUUUUCGCAAUGCUUGUUGCGGUUUU-- ......(((((...((((((((((((((..((((--------.((((........))))))))...)))))))...((((......)))).......))))))).)))))-- ( -25.80, z-score = -0.96, R) >droMoj3.scaffold_6680 17923965 93 - 24764193 CUCAUUAAAACAGCG---------AAGGCGUAUA--------CGUGGAU-CUG-CCCAACGUAAAAUUUCCAUAUUGAAAUUGAAUUUUCGCAAUGCGUGUUGCGGUUUUGC .....((((((.(((---------(..(((((((--------(((((..-...-.)).))))).((((((......))))))............))))).)))).)))))). ( -22.20, z-score = -0.71, R) >consensus CUCAUUAAAGCGAUCGGCGGAGGGGUGGGGG_____________U_ACCGCGAUUC__GGUUGCAAUUUCCAUAUGGAAAUUGAAUUUCCACCGGCCUUGAUGUGGUUUU__ ...............................................................(((((((......))))))).....((((..........))))...... ( -8.30 = -7.74 + -0.56)
| Location | 12,715,572 – 12,715,675 |
|---|---|
| Length | 103 |
| Sequences | 11 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 64.17 |
| Shannon entropy | 0.74438 |
| G+C content | 0.47657 |
| Mean single sequence MFE | -25.67 |
| Consensus MFE | -5.57 |
| Energy contribution | -5.83 |
| Covariance contribution | 0.26 |
| Combinations/Pair | 1.45 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.22 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.842061 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
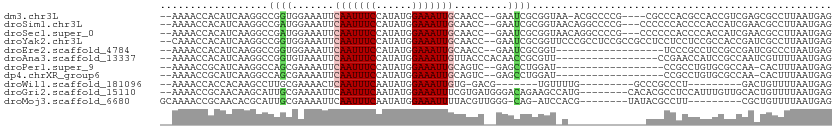
>dm3.chr3L 12715572 103 - 24543557 --AAAACCACAUCAAGGCCGGUGGAAAUUCAAUUUCCAUAUGGAAAUUGCAACC--GAAUCGCGGUAA-ACGCCCCG----CGCCCACGCCACCGUCGAGCGCCUUAAUGAG --.......(((.(((((((((((.....((((((((....)))))))).....--....(((((...-.....)))----))......))))))......))))).))).. ( -34.10, z-score = -3.06, R) >droSim1.chr3L 12099858 105 - 22553184 --AAAACCACAUCAAGGCCGAUGGAAAUUCAAUUUCCAUAUGGAAAUUGCAACC--GAAUCGCGGUAACAGGCCCCG---CCCCCCACCCCACCAUCGAACGCCUUAAUGAG --.......(((.(((((((((((.....((((((((....))))))))..(((--(.....))))....(((...)---))..........))))))...))))).))).. ( -31.90, z-score = -4.55, R) >droSec1.super_0 4900995 105 - 21120651 --AAAACCACAUCAAGGCCGAUGGAAAUUCAAUUUCCAUAUGGAAAUUGCAACC--GAAUCGCGGUAACAGGCCCCG---CCCCCCACCCCACCAUCGAACGCCUUAAUGAG --.......(((.(((((((((((.....((((((((....))))))))..(((--(.....))))....(((...)---))..........))))))...))))).))).. ( -31.90, z-score = -4.55, R) >droYak2.chr3L 12786490 108 - 24197627 --CAAACCACAUCAAGGCCGGUGGAAAUUCAAUUUCCAUAUGGAAAUUGCAACC--GAAUCGCGGUUCCCGCCUCCGCCGCCUCCUCCUCCGCCACCGAUCGCCUUAAUGAG --.......(((.(((((((((((.....((((((((....)))))))).....--.....(((((..........)))))...........))))))...))))).))).. ( -34.20, z-score = -3.64, R) >droEre2.scaffold_4784 12722291 90 - 25762168 --AAAACCACAUCAAGGCCGGUGGAAAUUCAAUUUCCAUAUGGAAAUUGCAACC--GAAUCGCGGU------------------UCCCGCCUCCGCCGAUCGCCCUAAUGAG --.......(((...((((((((((....((((((((....)))))))).....--.....((((.------------------..)))).)))))))...)))...))).. ( -29.50, z-score = -3.21, R) >droAna3.scaffold_13337 19983377 93 + 23293914 --AAAACCACAUCAAGGCCGGUGUAAAUUCAAUUUCCAUAUGGAAAUUGUUACCCACAACCGCGUU-----------------CCGAACCAUCCGCCAAUCGUUUUAAUGAG --.......(((.(((((.((.((((...((((((((....))))))))))))))......(((..-----------------..........))).....))))).))).. ( -18.60, z-score = -1.56, R) >droPer1.super_9 1803654 89 - 3637205 --AAAACCGCAUCAAGGCCAGCGAAAAUUCAAUUUCCAUAUGGAAAUUGCAGUC--GAGCCUGGAU------------------CCGCCUGUGCGCCAA-CACUUUAAUGAG --.....(((((..((((...(((.....((((((((....))))))))...))--).))))((..------------------...)).)))))....-............ ( -22.80, z-score = -1.47, R) >dp4.chrXR_group6 3508081 89 - 13314419 --AAAACCGCAUCAAGGCCAGCGAAAAUUCAAUUUCCAUAUGGAAAUUGCAGUC--GAGCCUGGAU------------------CCGCCUGUGCGCCAA-CACUUUAAUGAG --.....(((((..((((...(((.....((((((((....))))))))...))--).))))((..------------------...)).)))))....-............ ( -22.80, z-score = -1.47, R) >droWil1.scaffold_181096 11674647 84 + 12416693 --AAAACCACCACAAGCCUUGCGAAAACUCAAUUUCAAUAUGGAAAUUGUG-GACG-------UGUUUUG---------GCCCGCCCU---------GACUGUUUUAAUGAG --(((((..((((((.((....((((......)))).....))...)))))-)...-------.(((..(---------(....))..---------))).)))))...... ( -16.10, z-score = -0.45, R) >droGri2.scaffold_15110 7785130 102 + 24565398 --AAAACCGCAACAAGCAUUGCGAAAAUUCAAUUUCAAUAUGGAAAUUUCGUGAUGGGACAGAAGCCAUG--------CACACGCCUCCAUUUGUUGCACUGUUUUAAUGAG --(((((.(((((((((((..(((((.(((.((......)).))).)))))..)))(((..(..((...)--------)...)...))).))))))))...)))))...... ( -23.40, z-score = -0.93, R) >droMoj3.scaffold_6680 17923965 93 + 24764193 GCAAAACCGCAACACGCAUUGCGAAAAUUCAAUUUCAAUAUGGAAAUUUUACGUUGGG-CAG-AUCCACG--------UAUACGCCUU---------CGCUGUUUUAAUGAG ..(((((.((.....))...(((((.....((((((......)))))).(((((.(((-...-.))))))--------))......))---------))).)))))...... ( -17.10, z-score = 0.07, R) >consensus __AAAACCACAUCAAGGCCGGUGAAAAUUCAAUUUCCAUAUGGAAAUUGCAACC__GAACCGCGGU_A_____________CCCCCACCCCUCCGCCGACCGCUUUAAUGAG ..................((((.......(((((((......))))))).........)))).................................................. ( -5.57 = -5.83 + 0.26)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:21:52 2011