| Sequence ID | dm3.chr3L |
|---|---|
| Location | 12,707,647 – 12,707,746 |
| Length | 99 |
| Max. P | 0.824373 |

| Location | 12,707,647 – 12,707,746 |
|---|---|
| Length | 99 |
| Sequences | 9 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 73.36 |
| Shannon entropy | 0.51307 |
| G+C content | 0.46359 |
| Mean single sequence MFE | -29.33 |
| Consensus MFE | -16.89 |
| Energy contribution | -16.94 |
| Covariance contribution | 0.05 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.824373 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
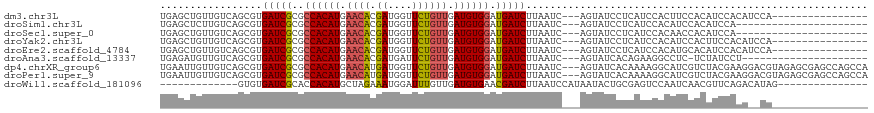
>dm3.chr3L 12707647 99 + 24543557 UGAGCUGUUGUCAGCGUGAUCGCGCCACAUGAACACGAUGGUUCUGUUGAUGUGGAUGAUCUUAAUC---AGUAUCCUCAUCCACUUCCACAUCCACAUCCA---------------- .((((((((((((((((....))))....)))...)))))))))(((.((.((((((((........---.......)))))))).)).)))..........---------------- ( -26.86, z-score = -0.78, R) >droSim1.chr3L 12091983 93 + 22553184 UGAGCUCUUGUCAGCGUGAUCGCGCCACAUGAACACGAUGGUUCUGUUGAUGUGGAUGAUCUUAAUC---AGUAUCCUCAUCCACAUCCACAUCCA---------------------- .(((((.((((((((((....))))....)))...))).)))))(((.(((((((((((........---.......))))))))))).)))....---------------------- ( -28.36, z-score = -1.70, R) >droSec1.super_0 4893041 93 + 21120651 UGAGCUGUUGUCAGCGUGAUCGCGCCACAUGAACACGAUGGUUCUGUUGAUGUGGAUGAUCUUAAUC---AGUAUCCUCAUCCACAACCACAUCCA---------------------- .((((((((((((((((....))))....)))...)))))))))(((.(.(((((((((........---.......))))))))).).)))....---------------------- ( -26.56, z-score = -0.93, R) >droYak2.chr3L 12778359 99 + 24197627 UGAGCUGUUGUCAGCGUGAUCGCGCCACAUGAACACGAUGGUUCUGUUGAUGUGGAUGAUCUUAAUC---AGUAUCCUCAUCCACAUCCACUUCCACAUCCA---------------- ...((((....))))(((.(((.......))).)))((((.....((.(((((((((((........---.......))))))))))).)).....))))..---------------- ( -30.26, z-score = -1.86, R) >droEre2.scaffold_4784 12712817 99 + 25762168 UGAGCUGUUGUCAGCGUGAUCGCGCCACAUGAACACGAUGGUUCUGUUGAUGUGGAUGAUCUUAAUC---AGUAUCCUCAUCCACAUGCACAUCCACAUCCA---------------- .((((((((((((((((....))))....)))...)))))))))(((..((((((((((........---.......))))))))))..)))..........---------------- ( -27.16, z-score = -0.21, R) >droAna3.scaffold_13337 19976137 93 - 23293914 UGAGAUGUUGUCAGCGUGAUCGCGCCACAUGAACACGAUGAUUCUGUUGAUGUGGAUGAUCUUAAUC---AGUAUCACAGAAGGCCUC-UCUAUCCU--------------------- .((((.((..((...(((((.((.((((((.((((.((....)))))).)))))).((((....)))---)))))))).))..)).))-))......--------------------- ( -28.20, z-score = -1.36, R) >dp4.chrXR_group6 3501000 115 + 13314419 UGAAUUGUUGUCAGCGUGAUCGCGCCACAUGAACAUGAUGGUUCUGUUGAUGUGGAUGAUCUUAAUC---AGUAUCACAAAAGGCAUCGUCUACGAAGGACGUAGAGCGAGCCAGCCA .............(((((((.((.((((((.((((.((....)))))).)))))).((((....)))---))))))))....(((.(((((((((.....)))))).)))))).)).. ( -39.20, z-score = -2.24, R) >droPer1.super_9 1796594 115 + 3637205 UGAAUUGUUGUCAGCGUGAUCGCGCCACAUGAACAUGAUGGUUCUGUUGAUGUGGAUGAUCUUAAUC---AGUAUCACAAAAGGCAUCGUCUACGAAGGACGUAGAGCGAGCCAGCCA .............(((((((.((.((((((.((((.((....)))))).)))))).((((....)))---))))))))....(((.(((((((((.....)))))).)))))).)).. ( -39.20, z-score = -2.24, R) >droWil1.scaffold_181096 833222 90 + 12416693 -------------GUGUGAUCGCACCACAUGCUAGAAAUGGAUUUGUUGAUGUGAACGAUCUUAAUCCAUAAUACUGCGAGUCCAAUCAACGUUCAGACAUAG--------------- -------------((.(((.((((.....)))......(((((((((.(.(((....(((....)))....)))).)))))))))......).))).))....--------------- ( -18.20, z-score = -0.09, R) >consensus UGAGCUGUUGUCAGCGUGAUCGCGCCACAUGAACACGAUGGUUCUGUUGAUGUGGAUGAUCUUAAUC___AGUAUCCUCAUCCACAUCCACAUCCACA__CA________________ .................(((((..((((((.((((.((....)))))).)))))).)))))......................................................... (-16.89 = -16.94 + 0.05)
| Location | 12,707,647 – 12,707,746 |
|---|---|
| Length | 99 |
| Sequences | 9 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 73.36 |
| Shannon entropy | 0.51307 |
| G+C content | 0.46359 |
| Mean single sequence MFE | -30.38 |
| Consensus MFE | -13.87 |
| Energy contribution | -14.02 |
| Covariance contribution | 0.15 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.596426 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 12707647 99 - 24543557 ----------------UGGAUGUGGAUGUGGAAGUGGAUGAGGAUACU---GAUUAAGAUCAUCCACAUCAACAGAACCAUCGUGUUCAUGUGGCGCGAUCACGCUGACAACAGCUCA ----------------.((.(((.((((((((.(..(((.((....))---.))).....).)))))))).)))...))((((((((.....))))))))...((((....))))... ( -31.20, z-score = -1.09, R) >droSim1.chr3L 12091983 93 - 22553184 ----------------------UGGAUGUGGAUGUGGAUGAGGAUACU---GAUUAAGAUCAUCCACAUCAACAGAACCAUCGUGUUCAUGUGGCGCGAUCACGCUGACAAGAGCUCA ----------------------(((.(((.(((((((((((.(((...---.)))....))))))))))).)))...)))..(.((((.(((((((......)))).))).)))).). ( -34.30, z-score = -2.72, R) >droSec1.super_0 4893041 93 - 21120651 ----------------------UGGAUGUGGUUGUGGAUGAGGAUACU---GAUUAAGAUCAUCCACAUCAACAGAACCAUCGUGUUCAUGUGGCGCGAUCACGCUGACAACAGCUCA ----------------------.....(((((((((((((.(((...(---(((....))))))).)))).(((((((......)))).)))..)))))))))((((....))))... ( -29.90, z-score = -1.27, R) >droYak2.chr3L 12778359 99 - 24197627 ----------------UGGAUGUGGAAGUGGAUGUGGAUGAGGAUACU---GAUUAAGAUCAUCCACAUCAACAGAACCAUCGUGUUCAUGUGGCGCGAUCACGCUGACAACAGCUCA ----------------.....((((..((((((((((((((.(((...---.)))....))))))))))).(((((((......)))).)))..)))..))))((((....))))... ( -33.70, z-score = -1.86, R) >droEre2.scaffold_4784 12712817 99 - 25762168 ----------------UGGAUGUGGAUGUGCAUGUGGAUGAGGAUACU---GAUUAAGAUCAUCCACAUCAACAGAACCAUCGUGUUCAUGUGGCGCGAUCACGCUGACAACAGCUCA ----------------.....((((.(((((((((((((((.(((...---.)))....))))))))))..(((((((......)))).))).))))).))))((((....))))... ( -33.80, z-score = -1.50, R) >droAna3.scaffold_13337 19976137 93 + 23293914 ---------------------AGGAUAGA-GAGGCCUUCUGUGAUACU---GAUUAAGAUCAUCCACAUCAACAGAAUCAUCGUGUUCAUGUGGCGCGAUCACGCUGACAACAUCUCA ---------------------......((-((.....((((((((..(---(((....)))).((((((.((((((....)).)))).))))))....))))))..)).....)))). ( -21.20, z-score = 0.45, R) >dp4.chrXR_group6 3501000 115 - 13314419 UGGCUGGCUCGCUCUACGUCCUUCGUAGACGAUGCCUUUUGUGAUACU---GAUUAAGAUCAUCCACAUCAACAGAACCAUCAUGUUCAUGUGGCGCGAUCACGCUGACAACAAUUCA .(((.((((((.((((((.....))))))))).)))....(((((..(---(((....)))).((((((.((((((....)).)))).))))))....))))))))............ ( -34.90, z-score = -2.58, R) >droPer1.super_9 1796594 115 - 3637205 UGGCUGGCUCGCUCUACGUCCUUCGUAGACGAUGCCUUUUGUGAUACU---GAUUAAGAUCAUCCACAUCAACAGAACCAUCAUGUUCAUGUGGCGCGAUCACGCUGACAACAAUUCA .(((.((((((.((((((.....))))))))).)))....(((((..(---(((....)))).((((((.((((((....)).)))).))))))....))))))))............ ( -34.90, z-score = -2.58, R) >droWil1.scaffold_181096 833222 90 - 12416693 ---------------CUAUGUCUGAACGUUGAUUGGACUCGCAGUAUUAUGGAUUAAGAUCGUUCACAUCAACAAAUCCAUUUCUAGCAUGUGGUGCGAUCACAC------------- ---------------............((.(((((.((.((((((...(((((((..(((.(....))))....))))))).....)).)))))).)))))))..------------- ( -19.50, z-score = -0.15, R) >consensus ________________UG__UGUGGAAGUGGAUGUGGAUGAGGAUACU___GAUUAAGAUCAUCCACAUCAACAGAACCAUCGUGUUCAUGUGGCGCGAUCACGCUGACAACAGCUCA .........................................................((((..((((((.((((((....)).)))).))))))...))))................. (-13.87 = -14.02 + 0.15)
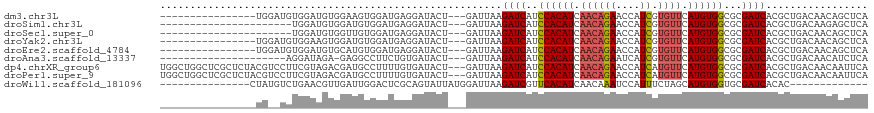

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:21:50 2011