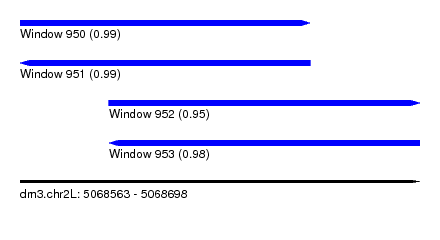
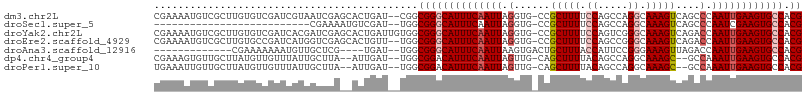
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 5,068,563 – 5,068,698 |
| Length | 135 |
| Max. P | 0.986272 |

| Location | 5,068,563 – 5,068,661 |
|---|---|
| Length | 98 |
| Sequences | 9 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 69.64 |
| Shannon entropy | 0.59290 |
| G+C content | 0.46931 |
| Mean single sequence MFE | -29.80 |
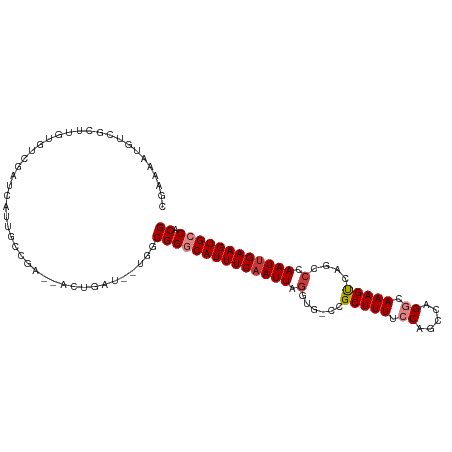
| Consensus MFE | -14.48 |
| Energy contribution | -14.33 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.23 |
| SVM RNA-class probability | 0.986272 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
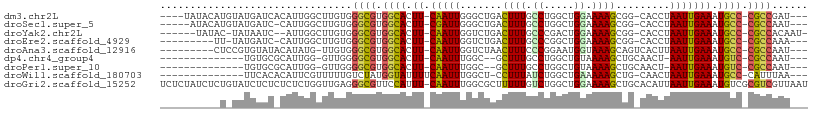
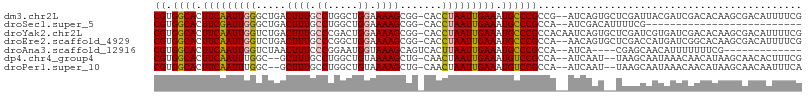
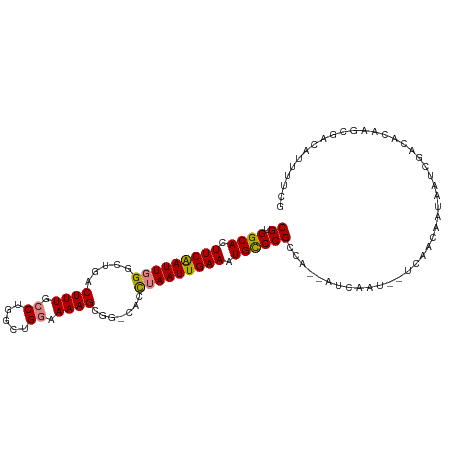
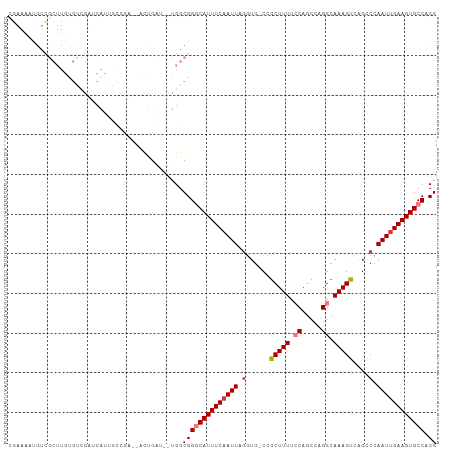
>dm3.chr2L 5068563 98 + 23011544 ----UAUACAUGUAUGAUCACAUUGGCUUGUGGGCGUGGCACUU-CAAUUGGGCUGACUUUGCCUGGCUGGAAAAGCGG-CACCUAAUUGAAAUGCC-CGCCGAU--- ----..............((((......))))((((.((((.((-(((((((((((.((((.((.....)).)))))))-).)).))))))).))))-))))...--- ( -39.30, z-score = -3.61, R) >droSec1.super_5 3155472 96 + 5866729 -----AUACAUGUAUGAUC-CAUUGGCUUGUGGGCGUGGCACUU-CGAUUGGGCUGACUUUGCCUGGCUGGAAAAGCGG-CACCUAAUUGAAAUGCC-CGCCAAU--- -----.((((.((......-.....)).))))((((.((((.((-(((((((((((.((((.((.....)).)))))))-).)).))))))).))))-))))...--- ( -36.80, z-score = -2.69, R) >droYak2.chr2L 11868077 95 - 22324452 ------UAUAC-UAUAAUC--AUUGGCUUGUGGGCGUGGCACUU-CAAUUGGUCUGACUUUGCCCGACUGGAAAAGCGG-CACCUAAUUGAAAUGCC-CGCCACAAU- ------.....-.......--......((((((.((.((((.((-(((((((((((.((((.((.....)).)))))))-.))).))))))).))))-)))))))).- ( -34.50, z-score = -3.77, R) >droEre2.scaffold_4929 5148425 91 + 26641161 ---------UU-UAUGAUC-CAUUGGCUUGUGGGCGUGGCACUU-CAAUUGGUCUGACUUUGCCCGGCUGGAAAAGCGG-CACCUAAUUGAAAUGCC-CGCCAAA--- ---------..-.......-(((......)))((((.((((.((-(((((((((((.((((.((.....)).)))))))-.))).))))))).))))-))))...--- ( -33.00, z-score = -2.12, R) >droAna3.scaffold_12916 13706195 93 + 16180835 ---------CUCCGUGUAUACAUAUG-UUGUGGGCGUGGCACUU-CAAUUGGUCUAACUUUCCCGGAAUGGUAAAGCAGUCACUUAAUUGAAAUGCC-CGCCAAU--- ---------.........((((....-.))))((((.((((.((-((((((((....((((.((.....)).)))).....)).)))))))).))))-))))...--- ( -26.60, z-score = -1.66, R) >dp4.chr4_group4 1588268 85 + 6586962 --------------UGUGCGCAUUGG-GUUGGGGCGUGGCACUU-CAAUUUGGC--GCUUUGCCUGGCUGUAAAAGCUGCAACU-AAUUGAAAUGUC-CGCCAAU--- --------------.(((.(((((.(-(((((((((.(((.((.-......)).--))).)))))((((.....)))).)))))-......))))).-)))....--- ( -26.30, z-score = 0.13, R) >droPer1.super_10 591774 85 + 3432795 --------------UGUGCGCAUUGG-GUUGGGGCGUGGCACUU-CAAUUUGGC--GCUUUGCCUGGCUGUAAAAGCUGCAACU-AAUUGAAAUGUC-CGCCAAU--- --------------.(((.(((((.(-(((((((((.(((.((.-......)).--))).)))))((((.....)))).)))))-......))))).-)))....--- ( -26.30, z-score = 0.13, R) >droWil1.scaffold_180703 1359543 88 + 3946847 --------------UUCACACAUUCGUUUUUGUCUAUGGUAUUUUCAAUUUGGCU-CCUUUAUCUGGCUGAAAAAGCUG-CAACUAAUUGAAAUGCC-CAUUUAA--- --------------.......................(((((((.(((((..((.-.((((.((.....)).))))..)-)....))))))))))))-.......--- ( -16.30, z-score = -1.33, R) >droGri2.scaffold_15252 11801610 107 + 17193109 UCUCUAUCUCUGUAUCUCUCUCUCUGGUUGAGGGCGUUCCAUUU-CAAUUUGGCGCUUUUUGUCUGGCUGGAAAAGCUGCACAUUAAUUGAAAUGUCGCGUCGUUAAU ..........................(((((.(((((..(((((-((((((((((((((((.........))))))).)).))..))))))))))..))))).))))) ( -29.10, z-score = -2.99, R) >consensus __________U_UAUGAUC_CAUUGGCUUGUGGGCGUGGCACUU_CAAUUGGGCUGACUUUGCCUGGCUGGAAAAGCGG_CACCUAAUUGAAAUGCC_CGCCAAU___ ................................((((.((((.((.(((((.......((((.((.....)).)))).........))))))).)))).))))...... (-14.48 = -14.33 + -0.14)

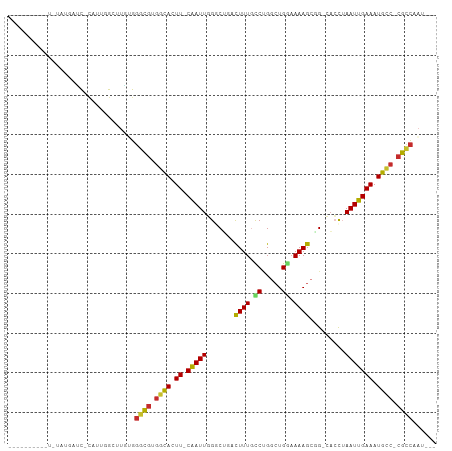
| Location | 5,068,563 – 5,068,661 |
|---|---|
| Length | 98 |
| Sequences | 9 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 69.64 |
| Shannon entropy | 0.59290 |
| G+C content | 0.46931 |
| Mean single sequence MFE | -27.67 |
| Consensus MFE | -11.59 |
| Energy contribution | -12.21 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.20 |
| SVM RNA-class probability | 0.985515 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 5068563 98 - 23011544 ---AUCGGCG-GGCAUUUCAAUUAGGUG-CCGCUUUUCCAGCCAGGCAAAGUCAGCCCAAUUG-AAGUGCCACGCCCACAAGCCAAUGUGAUCAUACAUGUAUA---- ---...((((-((((((((((((.(((.-..(((((.((.....)).)))))..))).)))))-))))))).))))((((......))))..............---- ( -38.10, z-score = -4.47, R) >droSec1.super_5 3155472 96 - 5866729 ---AUUGGCG-GGCAUUUCAAUUAGGUG-CCGCUUUUCCAGCCAGGCAAAGUCAGCCCAAUCG-AAGUGCCACGCCCACAAGCCAAUG-GAUCAUACAUGUAU----- ---...((((-((((((((.(((.(((.-..(((((.((.....)).)))))..))).))).)-))))))).)))).........(((-.......)))....----- ( -31.20, z-score = -2.19, R) >droYak2.chr2L 11868077 95 + 22324452 -AUUGUGGCG-GGCAUUUCAAUUAGGUG-CCGCUUUUCCAGUCGGGCAAAGUCAGACCAAUUG-AAGUGCCACGCCCACAAGCCAAU--GAUUAUA-GUAUA------ -.((((((((-((((((((((((.(((.-..((((((((....))).)))))...))))))))-))))))).)).))))))......--.......-.....------ ( -36.80, z-score = -4.28, R) >droEre2.scaffold_4929 5148425 91 - 26641161 ---UUUGGCG-GGCAUUUCAAUUAGGUG-CCGCUUUUCCAGCCGGGCAAAGUCAGACCAAUUG-AAGUGCCACGCCCACAAGCCAAUG-GAUCAUA-AA--------- ---...((((-((((((((((((.(((.-..((((((((....))).)))))...))))))))-))))))).))))............-.......-..--------- ( -33.50, z-score = -2.87, R) >droAna3.scaffold_12916 13706195 93 - 16180835 ---AUUGGCG-GGCAUUUCAAUUAAGUGACUGCUUUACCAUUCCGGGAAAGUUAGACCAAUUG-AAGUGCCACGCCCACAA-CAUAUGUAUACACGGAG--------- ---...((((-((((((((((((..((....(((((.((.....)).)))))...)).)))))-))))))).)))).....-.................--------- ( -28.70, z-score = -3.06, R) >dp4.chr4_group4 1588268 85 - 6586962 ---AUUGGCG-GACAUUUCAAUU-AGUUGCAGCUUUUACAGCCAGGCAAAGC--GCCAAAUUG-AAGUGCCACGCCCCAAC-CCAAUGCGCACA-------------- ---...((((-(.((((((((((-.......(((.....)))..(((.....--))).)))))-))))).).)))).....-............-------------- ( -22.80, z-score = -0.80, R) >droPer1.super_10 591774 85 - 3432795 ---AUUGGCG-GACAUUUCAAUU-AGUUGCAGCUUUUACAGCCAGGCAAAGC--GCCAAAUUG-AAGUGCCACGCCCCAAC-CCAAUGCGCACA-------------- ---...((((-(.((((((((((-.......(((.....)))..(((.....--))).)))))-))))).).)))).....-............-------------- ( -22.80, z-score = -0.80, R) >droWil1.scaffold_180703 1359543 88 - 3946847 ---UUAAAUG-GGCAUUUCAAUUAGUUG-CAGCUUUUUCAGCCAGAUAAAGG-AGCCAAAUUGAAAAUACCAUAGACAAAAACGAAUGUGUGAA-------------- ---....(((-(...((((((((....(-(..((((.((.....)).)))).-.))..))))))))...)))).....................-------------- ( -14.00, z-score = -0.03, R) >droGri2.scaffold_15252 11801610 107 - 17193109 AUUAACGACGCGACAUUUCAAUUAAUGUGCAGCUUUUCCAGCCAGACAAAAAGCGCCAAAUUG-AAAUGGAACGCCCUCAACCAGAGAGAGAGAUACAGAGAUAGAGA .........(((.((((((((((..((.((.((((((...........)))))))))))))))-)))))...))).(((.....)))..................... ( -21.10, z-score = -2.40, R) >consensus ___AUUGGCG_GGCAUUUCAAUUAGGUG_CAGCUUUUCCAGCCAGGCAAAGUCAGCCCAAUUG_AAGUGCCACGCCCACAAGCCAAUG_GAACAUA_A__________ ......((((.(.((((((((((........(((((.((.....)).)))))......))))).))))).).))))................................ (-11.59 = -12.21 + 0.62)

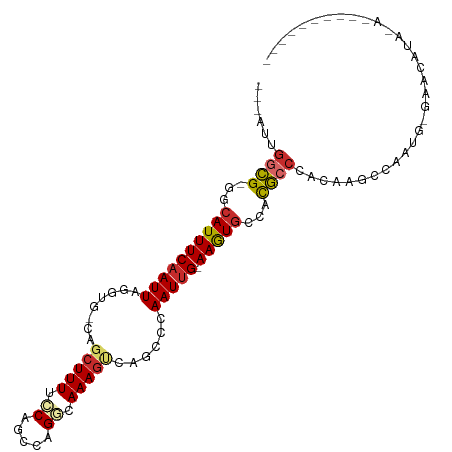
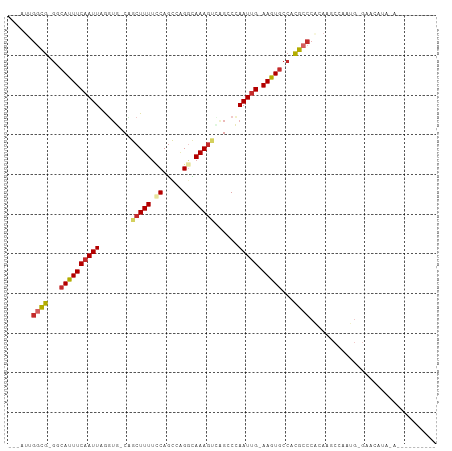
| Location | 5,068,593 – 5,068,698 |
|---|---|
| Length | 105 |
| Sequences | 7 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 71.30 |
| Shannon entropy | 0.56587 |
| G+C content | 0.47898 |
| Mean single sequence MFE | -26.69 |
| Consensus MFE | -16.17 |
| Energy contribution | -16.30 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.51 |
| SVM RNA-class probability | 0.945579 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 5068593 105 + 23011544 CGUGGCACUUCAAUUGGGCUGACUUUGCCUGGCUGGAAAAGCGG-CACCUAAUUGAAAUGCCCGCCG--AUCAGUGCUCGAUUACGAUCGACACAAGCGACAUUUUCG (((((((.(((((((((((((.((((.((.....)).)))))))-).)).))))))).)))).....--....(((.(((((....))))))))..)))......... ( -36.00, z-score = -2.64, R) >droSec1.super_5 3155500 79 + 5866729 CGUGGCACUUCGAUUGGGCUGACUUUGCCUGGCUGGAAAAGCGG-CACCUAAUUGAAAUGCCCGCCA--AUCGACAUUUUCG-------------------------- ((.((((.(((((((((((((.((((.((.....)).)))))))-).)).))))))).))))))...--.............-------------------------- ( -27.50, z-score = -2.54, R) >droYak2.chr2L 11868102 107 - 22324452 CGUGGCACUUCAAUUGGUCUGACUUUGCCCGACUGGAAAAGCGG-CACCUAAUUGAAAUGCCCGCCACAAUCAGUGCUCGAUCGUGAUCGACACAAGCGACAUUUUCG (((((((.(((((((((((((.((((.((.....)).)))))))-.))).))))))).))))...........(((.(((((....))))))))..)))......... ( -33.40, z-score = -2.16, R) >droEre2.scaffold_4929 5148448 105 + 26641161 CGUGGCACUUCAAUUGGUCUGACUUUGCCCGGCUGGAAAAGCGG-CACCUAAUUGAAAUGCCCGCCA--AACAGUGCUCGACCAUGAUCGGCACAAGCGACAUUUUCG (((((((.(((((((((((((.((((.((.....)).)))))))-.))).))))))).)))).....--....((((.(((......)))))))..)))......... ( -33.50, z-score = -1.60, R) >droAna3.scaffold_12916 13706219 89 + 16180835 CGUGGCACUUCAAUUGGUCUAACUUUCCCGGAAUGGUAAAGCAGUCACUUAAUUGAAAUGCCCGCCA--AUCA----CGAGCAACAUUUUUUUCG------------- .(.((((.((((((((((....((((.((.....)).)))).....)).)))))))).)))))(((.--....----.).)).............------------- ( -17.40, z-score = -0.37, R) >dp4.chr4_group4 1588287 101 + 6586962 CGUGGCACUUCAAUUUGGC--GCUUUGCCUGGCUGUAAAAGCUG-CAACUAAUUGAAAUGUCCGCCA--AUCAAU--UAAGCAAUAAACAACAUAAGCAACACUUUCG .((((((.(((((((..((--(((((..(.....)..))))).)-)....))))))).)).))))..--......--............................... ( -19.50, z-score = 0.16, R) >droPer1.super_10 591793 101 + 3432795 CGUGGCACUUCAAUUUGGC--GCUUUGCCUGGCUGUAAAAGCUG-CAACUAAUUGAAAUGUCCGCCA--AUCAAU--UAAGCAAUAAACAACAUAAGCAACAAUUUCA .((((((.(((((((..((--(((((..(.....)..))))).)-)....))))))).)).))))..--......--............................... ( -19.50, z-score = 0.27, R) >consensus CGUGGCACUUCAAUUGGGCUGACUUUGCCUGGCUGGAAAAGCGG_CACCUAAUUGAAAUGCCCGCCA__AUCAAU__UCAACAAUAAUCGACACAAGCGACAUUUUCG ((.((((.(((((((((.....((((.((.....)).)))).......))))))))).))))))............................................ (-16.17 = -16.30 + 0.13)
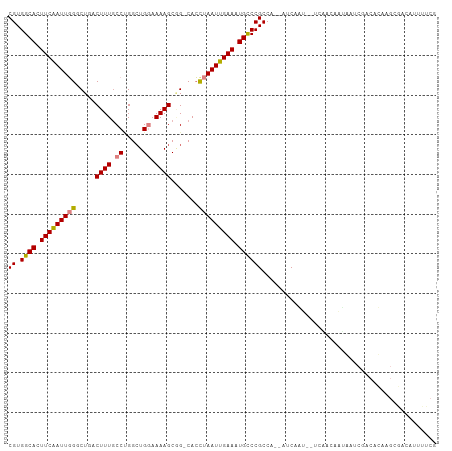
| Location | 5,068,593 – 5,068,698 |
|---|---|
| Length | 105 |
| Sequences | 7 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 71.30 |
| Shannon entropy | 0.56587 |
| G+C content | 0.47898 |
| Mean single sequence MFE | -31.39 |
| Consensus MFE | -17.07 |
| Energy contribution | -17.73 |
| Covariance contribution | 0.65 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.96 |
| SVM RNA-class probability | 0.976737 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 5068593 105 - 23011544 CGAAAAUGUCGCUUGUGUCGAUCGUAAUCGAGCACUGAU--CGGCGGGCAUUUCAAUUAGGUG-CCGCUUUUCCAGCCAGGCAAAGUCAGCCCAAUUGAAGUGCCACG ......(((((...((((((((....))))).)))....--)))))((((((((((((.(((.-..(((((.((.....)).)))))..))).))))))))))))... ( -39.40, z-score = -3.22, R) >droSec1.super_5 3155500 79 - 5866729 --------------------------CGAAAAUGUCGAU--UGGCGGGCAUUUCAAUUAGGUG-CCGCUUUUCCAGCCAGGCAAAGUCAGCCCAAUCGAAGUGCCACG --------------------------......((((...--.))))((((((((.(((.(((.-..(((((.((.....)).)))))..))).))).))))))))... ( -24.80, z-score = -1.57, R) >droYak2.chr2L 11868102 107 + 22324452 CGAAAAUGUCGCUUGUGUCGAUCACGAUCGAGCACUGAUUGUGGCGGGCAUUUCAAUUAGGUG-CCGCUUUUCCAGUCGGGCAAAGUCAGACCAAUUGAAGUGCCACG ......((((((..((((((((....))))).))).....))))))((((((((((((.(((.-..((((((((....))).)))))...)))))))))))))))... ( -40.00, z-score = -2.74, R) >droEre2.scaffold_4929 5148448 105 - 26641161 CGAAAAUGUCGCUUGUGCCGAUCAUGGUCGAGCACUGUU--UGGCGGGCAUUUCAAUUAGGUG-CCGCUUUUCCAGCCGGGCAAAGUCAGACCAAUUGAAGUGCCACG .........((((.((((((((....)))).))))....--.))))((((((((((((.(((.-..((((((((....))).)))))...)))))))))))))))... ( -41.30, z-score = -2.85, R) >droAna3.scaffold_12916 13706219 89 - 16180835 -------------CGAAAAAAAUGUUGCUCG----UGAU--UGGCGGGCAUUUCAAUUAAGUGACUGCUUUACCAUUCCGGGAAAGUUAGACCAAUUGAAGUGCCACG -------------((..((..(((.....))----)..)--)..))((((((((((((..((....(((((.((.....)).)))))...)).))))))))))))... ( -22.90, z-score = -1.15, R) >dp4.chr4_group4 1588287 101 - 6586962 CGAAAGUGUUGCUUAUGUUGUUUAUUGCUUA--AUUGAU--UGGCGGACAUUUCAAUUAGUUG-CAGCUUUUACAGCCAGGCAAAGC--GCCAAAUUGAAGUGCCACG .....(((..((((..(((((.....(((((--((((((--(((........)))))))))))-.)))....)))))..(((.....--)))......))))..))). ( -27.20, z-score = -0.83, R) >droPer1.super_10 591793 101 - 3432795 UGAAAUUGUUGCUUAUGUUGUUUAUUGCUUA--AUUGAU--UGGCGGACAUUUCAAUUAGUUG-CAGCUUUUACAGCCAGGCAAAGC--GCCAAAUUGAAGUGCCACG ........((((((..(((((.....(((((--((((((--(((........)))))))))))-.)))....))))).)))))).((--((.........)))).... ( -24.10, z-score = 0.02, R) >consensus CGAAAAUGUCGCUUGUGUCGAUCAUUGCCGA__ACUGAU__UGGCGGGCAUUUCAAUUAGGUG_CCGCUUUUCCAGCCAGGCAAAGUCAGCCCAAUUGAAGUGCCACG ............................................((((((((((((((.(......(((((.((.....)).)))))....).)))))))))))).)) (-17.07 = -17.73 + 0.65)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:17:57 2011