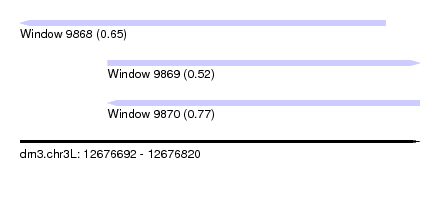
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 12,676,692 – 12,676,820 |
| Length | 128 |
| Max. P | 0.767928 |

| Location | 12,676,692 – 12,676,809 |
|---|---|
| Length | 117 |
| Sequences | 12 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 76.68 |
| Shannon entropy | 0.51136 |
| G+C content | 0.37079 |
| Mean single sequence MFE | -22.39 |
| Consensus MFE | -12.08 |
| Energy contribution | -11.96 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.652958 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 12676692 117 - 24543557 AAAUCCGCUUACAAUAAUUAAAAAGUUGC-AAAUUUAAGCAUUAACUCGGCGAGUGCGCAAAUAUGGCCAAAACGUAAGCGGAUUUCGCAUUAACUGCAGACAAUCGGCAAAACAAAC ((((((((((((..............(((-........))).......(((.(...........).))).....)))))))))))).(((.....))).................... ( -26.00, z-score = -1.50, R) >droSim1.chr3L 12062736 117 - 22553184 AAAUCCGCUUACAAUAAUUAAAAAGUUGC-AAAUUUAAGCAUUAACUCGGCGAGUGCGCAAAUAUGGCCAAAACGUAAGCGGAUUUCGCAUUAACUGCAGACAAUCGGCAAAACAAAC ((((((((((((..............(((-........))).......(((.(...........).))).....)))))))))))).(((.....))).................... ( -26.00, z-score = -1.50, R) >droSec1.super_0 4862581 117 - 21120651 AAAUCCGCUUACAAUAAUUAAAAAGUUGC-AAAUUUAAGCAUUAACUCGGCGAGUGCGUAAAUAUGGCCAAAACGUAAGCGGAUUUCGCAUUAACUGCAGACAAUCGGCAAAACAAAC ((((((((((((..............(((-........))).......(((.(...........).))).....)))))))))))).(((.....))).................... ( -26.00, z-score = -1.72, R) >droYak2.chr3L 12745898 117 - 24197627 AAAUCCGCUUACAAUAAUUAAAAAGUUGC-AAAUUUAAGUAUUAACUCGGCGAGUGCGCAGAUAUGGCCAAAACGUAAGCGGAUUUCGCAUUAACUGCAGACAAUCGGCAAAACAAAC ((((((((((((((((.(((((.......-...))))).)))).....(((.(...........).))).....)))))))))))).(((.....))).................... ( -24.20, z-score = -0.88, R) >droEre2.scaffold_4784 12681926 117 - 25762168 AAAUCCGCUUACAAUAAUUAAAAAGUUGC-AAAUUUAAGUAUUAACUCGGCGAGUGCGCAAAUAUGGCCAAAACGUAAGCGGAUUUCGCAUUAACUGCAGACAAUCGGCAAAACAAAC ((((((((((((((((.(((((.......-...))))).)))).....(((.(...........).))).....)))))))))))).(((.....))).................... ( -24.20, z-score = -1.10, R) >droAna3.scaffold_13337 19945308 116 + 23293914 GAAUCCGCUUACAAUAAUUAAAAAGUUGC-AAAUUUAAGUAUUAACUCGGCGAAUGCGCAAAUAUGGC-CAACGGUAAGCGGAUUUCGAAUUAACUGCAGACAAUCGGCAAAACAAAC ((((((((((((.............((((-.......(((....)))..((....)))))).......-.....)))))))))))).........(((.(.....).)))........ ( -23.91, z-score = -1.31, R) >dp4.chrXR_group6 3467354 117 - 13314419 GAAGCCGCUUACAAUAAUUAAAAAGUUGC-AAAUUUAAGUAUUAACUCUGCGAAUGCGCAAAUAUGGCCAGACCCUAAGCGGAUUUCGCAUUAACUGCAGACAAUCGGCAAAACAAAC ...((((((((((((.........)))).-.....)))))......((((((((((((.((((.(.((..........)).)))))))))))...)))))).....)))......... ( -22.00, z-score = -0.29, R) >droPer1.super_9 1762860 117 - 3637205 GAAGCCGCUUACAAUAAUUAAAAAGUUGC-AAAUUUAAGUAUUAACUCUGCGAAUGCGCAAAUAUGGCCAGACCCUAAGCGGAUUUCGCAUUAACUGCAGACAAUCGGCAAAACAAAC ...((((((((((((.........)))).-.....)))))......((((((((((((.((((.(.((..........)).)))))))))))...)))))).....)))......... ( -22.00, z-score = -0.29, R) >droWil1.scaffold_181096 11515855 101 + 12416693 AAAUCGGCUUAGAAUAAUUAAAAAUUAGG-GAAAUU--------------CGAUUGAUUGAAUACAACGUAAA-AUAAUCGGAUUUGGCCUUAGUUGCAGACAAUAACCAAAACAAA- .....((.(((...........(((((((-(.((((--------------((((((.((............))-.))))))))))...))))))))........)))))........- ( -16.41, z-score = -1.23, R) >droVir3.scaffold_13049 6093597 99 + 25233164 AAAUCCGCUUAGAAUGAUUAAAAAGUUGC-AAAUUUAAAGUAUUACCGUGCGAACGCACAAAUAUGGCC--AGCAUAAGCGGAUUU---CACAGACCCGAAAAAC------------- (((((((((((...((..........(((-.........))).....((((....)))).........)--)...)))))))))))---................------------- ( -21.00, z-score = -2.23, R) >droMoj3.scaffold_6680 17878375 99 + 24764193 AAAUCCGCUUAGAAUGAUUAAAAAGUUGC-AAAUUUAAGGUAUUACCGUGCGAAUGCACAAAUAUGGCC--AACAUAAUCGGAUUU---UCGAGACCCGAAAUUC------------- .((((((.(((...((.............-........((.....))((((....)))).........)--)...))).))))))(---(((.....))))....------------- ( -16.50, z-score = -0.08, R) >droGri2.scaffold_15110 7738990 102 + 24565398 AAAUCCGCUUAAAAUGAUUAAAAAGUUGCAAAAUUUAAAGUAUUAACGUGCGAACGCACAAAUAUGGCC--AACAUAAGCGGAUUU---CACAGUCCCGAAAAAAUU----------- (((((((((((...........(((((....)))))...........((((....))))..........--....)))))))))))---..................----------- ( -20.50, z-score = -2.23, R) >consensus AAAUCCGCUUACAAUAAUUAAAAAGUUGC_AAAUUUAAGUAUUAACUCGGCGAAUGCGCAAAUAUGGCCAAAACGUAAGCGGAUUUCGCAUUAACUGCAGACAAUCGGCAAAACAAAC (((((((((((...........(((((....)))))...................((.((....)))).......)))))))))))................................ (-12.08 = -11.96 + -0.12)



| Location | 12,676,720 – 12,676,820 |
|---|---|
| Length | 100 |
| Sequences | 12 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 74.72 |
| Shannon entropy | 0.51923 |
| G+C content | 0.39005 |
| Mean single sequence MFE | -23.56 |
| Consensus MFE | -10.02 |
| Energy contribution | -10.47 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.522169 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 12676720 100 + 24543557 UGCGAAAUCCGCUUACGUUUUGGCCAUAUUUGCGCACUCGCCGAGUUAAUGCUUAAAUUU-GCAACUUUUUAAUUAUUGUAAGCGGAUUUGCCAUCGAAAG----------------- .((.(((((((((((((.((((((...............))))))....(((........-))).............))))))))))))))).........----------------- ( -27.26, z-score = -2.11, R) >droSim1.chr3L 12062764 100 + 22553184 UGCGAAAUCCGCUUACGUUUUGGCCAUAUUUGCGCACUCGCCGAGUUAAUGCUUAAAUUU-GCAACUUUUUAAUUAUUGUAAGCGGAUUUGCCUUCGAAAG----------------- .((.(((((((((((((.((((((...............))))))....(((........-))).............))))))))))))))).........----------------- ( -27.26, z-score = -2.20, R) >droSec1.super_0 4862609 100 + 21120651 UGCGAAAUCCGCUUACGUUUUGGCCAUAUUUACGCACUCGCCGAGUUAAUGCUUAAAUUU-GCAACUUUUUAAUUAUUGUAAGCGGAUUUGCCUUCGAAAG----------------- .((.(((((((((((((.((((((...............))))))....(((........-))).............))))))))))))))).........----------------- ( -27.26, z-score = -2.80, R) >droYak2.chr3L 12745926 100 + 24197627 UGCGAAAUCCGCUUACGUUUUGGCCAUAUCUGCGCACUCGCCGAGUUAAUACUUAAAUUU-GCAACUUUUUAAUUAUUGUAAGCGGAUUUCCCUUCGAAAG----------------- ...(((((((((((((..((((((...............))))))..((((.(((((...-.......))))).)))))))))))))))))..........----------------- ( -26.36, z-score = -3.01, R) >droEre2.scaffold_4784 12681954 100 + 25762168 UGCGAAAUCCGCUUACGUUUUGGCCAUAUUUGCGCACUCGCCGAGUUAAUACUUAAAUUU-GCAACUUUUUAAUUAUUGUAAGCGGAUUUCCCUUCGAAAG----------------- ...((((((((((((((............((((((....)).((((....))))......-))))............))))))))))))))..........----------------- ( -26.85, z-score = -3.18, R) >droAna3.scaffold_13337 19945336 101 - 23293914 UUCGAAAUCCGCUUACCGU-UGGCCAUAUUUGCGCAUUCGCCGAGUUAAUACUUAAAUUU-GCAACUUUUUAAUUAUUGUAAGCGGAUUCCCUUUUUCGGGAC--------------- ......((((((((((.((-.........((((((....)).((((....))))......-))))..........)).))))))))))((((......)))).--------------- ( -25.31, z-score = -2.17, R) >dp4.chrXR_group6 3467382 111 + 13314419 UGCGAAAUCCGCUUAGGGUCUGGCCAUAUUUGCGCAUUCGCAGAGUUAAUACUUAAAUUU-GCAACUUUUUAAUUAUUGUAAGCGGCUUCUCCUUCUCCUGCUCCU------CGGGGA .(((.....)))...(((...((((...((((((....)))))).............(((-((((...........))))))).))))...)))(((((.......------.))))) ( -25.00, z-score = 0.36, R) >droPer1.super_9 1762888 117 + 3637205 UGCGAAAUCCGCUUAGGGUCUGGCCAUAUUUGCGCAUUCGCAGAGUUAAUACUUAAAUUU-GCAACUUUUUAAUUAUUGUAAGCGGCUUCUCCUUCUCCUGCUCCUGCUCCUCGGGGA .(((.....)))...(((...((((...((((((....)))))).............(((-((((...........))))))).))))...)))(((((.((....)).....))))) ( -25.70, z-score = 0.71, R) >droWil1.scaffold_181096 11515882 85 - 12416693 GGCCAAAUCCGAUUAUUUU------ACGUU---GUAUUCA-------AUCAAUCGAAUUUCCCUAAUUUUUAAUUAUUCUAAGCCGAUUUUCUCUGGAACG----------------- .......(((((.(((...------.....---))).)).-------...(((((..((....((((.....))))....))..)))))......)))...----------------- ( -6.50, z-score = 1.25, R) >droVir3.scaffold_13049 6093609 96 - 25233164 UGUGAAAUCCGCUUAUGCU--GGCCAUAUUUGUGCGUUCGCACGGUAAUACUUUAAAUUU-GCAACUUUUUAAUCAUUCUAAGCGGAUUUCUAGGCGAA------------------- ...((((((((((((.(.(--((.......(((((....)))))((((..........))-))..........))).).))))))))))))........------------------- ( -24.90, z-score = -1.96, R) >droMoj3.scaffold_6680 17878387 87 - 24764193 UCGAAAAUCCGAUUAUGUU--GGCCAUAUUUGUGCAUUCGCACGGUAAUACCUUAAAUUU-GCAACUUUUUAAUCAUUCUAAGCGGAUUU---------------------------- ....(((((((((((.(((--(.........((((....))))((.....))........-.))))....))))).........))))))---------------------------- ( -15.20, z-score = -0.56, R) >droGri2.scaffold_15110 7739004 97 - 24565398 UGUGAAAUCCGCUUAUGUU--GGCCAUAUUUGUGCGUUCGCACGUUAAUACUUUAAAUUUUGCAACUUUUUAAUCAUUUUAAGCGGAUUUCUAGGCGAU------------------- ...((((((((((((...(--(((((....)).))(((.(((..((((....))))....))))))........))...))))))))))))........------------------- ( -25.10, z-score = -2.54, R) >consensus UGCGAAAUCCGCUUACGUU_UGGCCAUAUUUGCGCAUUCGCCGAGUUAAUACUUAAAUUU_GCAACUUUUUAAUUAUUGUAAGCGGAUUUCCCUUCGAAAG_________________ .....((((((((((................(((....)))...........(((((...........)))))......))))))))))............................. (-10.02 = -10.47 + 0.45)

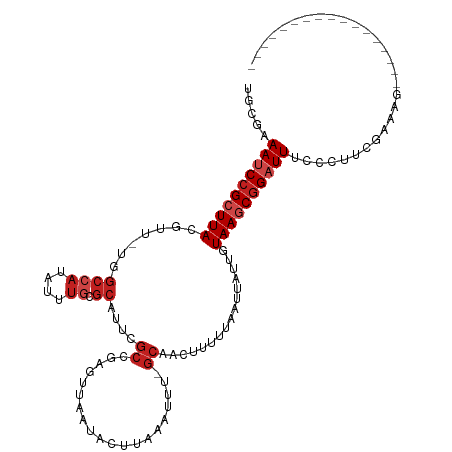
| Location | 12,676,720 – 12,676,820 |
|---|---|
| Length | 100 |
| Sequences | 12 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 74.72 |
| Shannon entropy | 0.51923 |
| G+C content | 0.39005 |
| Mean single sequence MFE | -23.68 |
| Consensus MFE | -11.89 |
| Energy contribution | -12.30 |
| Covariance contribution | 0.41 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.767928 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 12676720 100 - 24543557 -----------------CUUUCGAUGGCAAAUCCGCUUACAAUAAUUAAAAAGUUGC-AAAUUUAAGCAUUAACUCGGCGAGUGCGCAAAUAUGGCCAAAACGUAAGCGGAUUUCGCA -----------------.........((((((((((((((..............(((-........))).......(((.(...........).))).....)))))))))))).)). ( -27.60, z-score = -2.17, R) >droSim1.chr3L 12062764 100 - 22553184 -----------------CUUUCGAAGGCAAAUCCGCUUACAAUAAUUAAAAAGUUGC-AAAUUUAAGCAUUAACUCGGCGAGUGCGCAAAUAUGGCCAAAACGUAAGCGGAUUUCGCA -----------------.........((((((((((((((..............(((-........))).......(((.(...........).))).....)))))))))))).)). ( -27.60, z-score = -2.33, R) >droSec1.super_0 4862609 100 - 21120651 -----------------CUUUCGAAGGCAAAUCCGCUUACAAUAAUUAAAAAGUUGC-AAAUUUAAGCAUUAACUCGGCGAGUGCGUAAAUAUGGCCAAAACGUAAGCGGAUUUCGCA -----------------.........((((((((((((((..............(((-........))).......(((.(...........).))).....)))))))))))).)). ( -27.60, z-score = -2.62, R) >droYak2.chr3L 12745926 100 - 24197627 -----------------CUUUCGAAGGGAAAUCCGCUUACAAUAAUUAAAAAGUUGC-AAAUUUAAGUAUUAACUCGGCGAGUGCGCAGAUAUGGCCAAAACGUAAGCGGAUUUCGCA -----------------........(.(((((((((((((((((.(((((.......-...))))).)))).....(((.(...........).))).....))))))))))))).). ( -28.40, z-score = -2.59, R) >droEre2.scaffold_4784 12681954 100 - 25762168 -----------------CUUUCGAAGGGAAAUCCGCUUACAAUAAUUAAAAAGUUGC-AAAUUUAAGUAUUAACUCGGCGAGUGCGCAAAUAUGGCCAAAACGUAAGCGGAUUUCGCA -----------------........(.(((((((((((((((((.(((((.......-...))))).)))).....(((.(...........).))).....))))))))))))).). ( -28.40, z-score = -2.84, R) >droAna3.scaffold_13337 19945336 101 + 23293914 ---------------GUCCCGAAAAAGGGAAUCCGCUUACAAUAAUUAAAAAGUUGC-AAAUUUAAGUAUUAACUCGGCGAAUGCGCAAAUAUGGCCA-ACGGUAAGCGGAUUUCGAA ---------------.((((......))))((((((((((.............((((-.......(((....)))..((....)))))).........-...))))))))))...... ( -26.31, z-score = -1.92, R) >dp4.chrXR_group6 3467382 111 - 13314419 UCCCCG------AGGAGCAGGAGAAGGAGAAGCCGCUUACAAUAAUUAAAAAGUUGC-AAAUUUAAGUAUUAACUCUGCGAAUGCGCAAAUAUGGCCAGACCCUAAGCGGAUUUCGCA ((.((.------.......)).))....(((((((((((..............((((-..((((.((........))..))))..))))..............))))))).))))... ( -20.59, z-score = 1.18, R) >droPer1.super_9 1762888 117 - 3637205 UCCCCGAGGAGCAGGAGCAGGAGAAGGAGAAGCCGCUUACAAUAAUUAAAAAGUUGC-AAAUUUAAGUAUUAACUCUGCGAAUGCGCAAAUAUGGCCAGACCCUAAGCGGAUUUCGCA (((....)))((....))..........(((((((((((..............((((-..((((.((........))..))))..))))..............))))))).))))... ( -20.89, z-score = 1.86, R) >droWil1.scaffold_181096 11515882 85 + 12416693 -----------------CGUUCCAGAGAAAAUCGGCUUAGAAUAAUUAAAAAUUAGGGAAAUUCGAUUGAU-------UGAAUAC---AACGU------AAAAUAAUCGGAUUUGGCC -----------------................((((.....((((.....))))...(((((((((((.(-------(......---.....------.)).))))))))))))))) ( -10.90, z-score = 0.69, R) >droVir3.scaffold_13049 6093609 96 + 25233164 -------------------UUCGCCUAGAAAUCCGCUUAGAAUGAUUAAAAAGUUGC-AAAUUUAAAGUAUUACCGUGCGAACGCACAAAUAUGGCC--AGCAUAAGCGGAUUUCACA -------------------........((((((((((((...((..........(((-.........))).....((((....)))).........)--)...))))))))))))... ( -25.70, z-score = -3.13, R) >droMoj3.scaffold_6680 17878387 87 + 24764193 ----------------------------AAAUCCGCUUAGAAUGAUUAAAAAGUUGC-AAAUUUAAGGUAUUACCGUGCGAAUGCACAAAUAUGGCC--AACAUAAUCGGAUUUUCGA ----------------------------(((((((.(((...((.............-........((.....))((((....)))).........)--)...))).))))))).... ( -15.00, z-score = -0.19, R) >droGri2.scaffold_15110 7739004 97 + 24565398 -------------------AUCGCCUAGAAAUCCGCUUAAAAUGAUUAAAAAGUUGCAAAAUUUAAAGUAUUAACGUGCGAACGCACAAAUAUGGCC--AACAUAAGCGGAUUUCACA -------------------........((((((((((((...........(((((....)))))...........((((....))))..........--....))))))))))))... ( -25.20, z-score = -3.63, R) >consensus _________________CUUUCGAAGGGAAAUCCGCUUACAAUAAUUAAAAAGUUGC_AAAUUUAAGUAUUAACUCGGCGAAUGCGCAAAUAUGGCCA_AACGUAAGCGGAUUUCGCA ............................(((((((((((...........(((((....))))).............((....))..................))))))))))).... (-11.89 = -12.30 + 0.41)

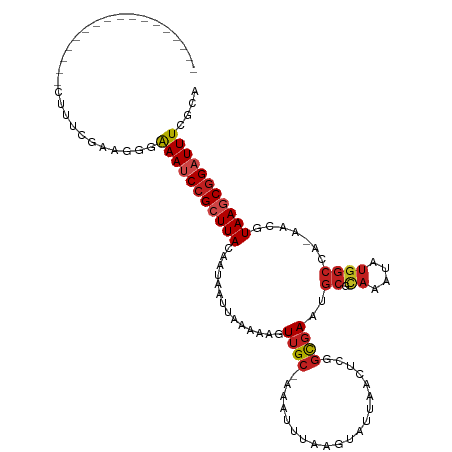

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:21:47 2011