
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 12,667,114 – 12,667,233 |
| Length | 119 |
| Max. P | 0.576852 |

| Location | 12,667,114 – 12,667,233 |
|---|---|
| Length | 119 |
| Sequences | 8 |
| Columns | 129 |
| Reading direction | reverse |
| Mean pairwise identity | 71.49 |
| Shannon entropy | 0.53897 |
| G+C content | 0.44266 |
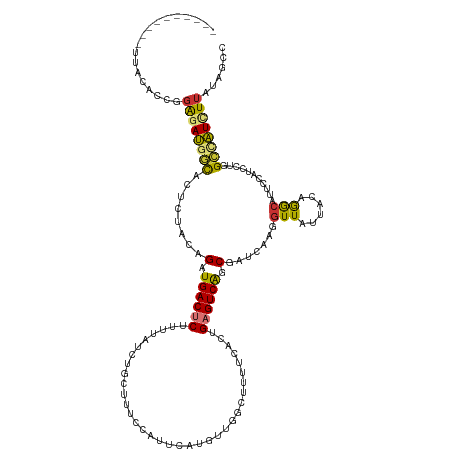
| Mean single sequence MFE | -29.64 |
| Consensus MFE | -13.08 |
| Energy contribution | -12.66 |
| Covariance contribution | -0.42 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.576852 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
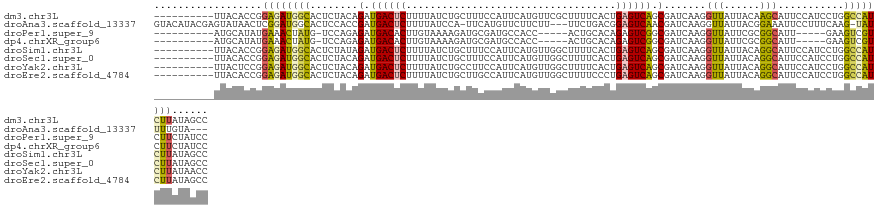
>dm3.chr3L 12667114 119 - 24543557 ----------UUACACCGGAGAUGGCACUCUACAGAUGACUCUUUUAUCUGCUUUCCAUUCAUGUUCGCUUUUCACUGAGUCAGCGAUCAAGGUUAUUACAAGCAUUCCAUCCUGGCCAUCUUAUAGCC ----------........((((((((.......((((((.....))))))((((.....((.((.(((((.(((...)))..))))).)).)).......))))...........))))))))...... ( -26.00, z-score = -1.18, R) >droAna3.scaffold_13337 19936007 121 + 23293914 GUACAUACGAGUAUAACUCGGAUGGCACUCCACCGAUGACUCUUUUAUCCA-UUCAUGUUCUUCUU---UUCUGACGGAGUCAACGAUCAAGGUUAUUACGGAAAUUCCUUUCAAG-UAUUUUGUA--- ((((...((((.....))))((((((.((....((.(((((((.(((....-..............---...))).))))))).))....))))))))..((.....))......)-)))......--- ( -21.50, z-score = 0.52, R) >droPer1.super_9 1753074 108 - 3637205 ----------AUGCAUAUGAAACUAUG-UCCAGAGAUGACACUUGUAAAAGAUGCGAUGCCACC-----ACUGCACAGAGUCGGCGAUCAAGGUUAUUCGCGGCAUU-----GAAGUCGUCUUCUAUCC ----------..(((((......))))-)...((((((((.(((....)))...(((((((.(.-----(((......))).)((((..........))))))))))-----)..))))))))...... ( -28.60, z-score = -0.75, R) >dp4.chrXR_group6 3457479 108 - 13314419 ----------AUGCAUAUGAAACUAUG-UCCAGAGAUGACACUUGUAAAAGAUGCGAUGCCACC-----ACUGCACAGAGUCGGCGAUCAAGGUUAUUCGCGGCAUU-----GAAGUCGUCUUCUAUCC ----------..(((((......))))-)...((((((((.(((....)))...(((((((.(.-----(((......))).)((((..........))))))))))-----)..))))))))...... ( -28.60, z-score = -0.75, R) >droSim1.chr3L 12053103 119 - 22553184 ----------UUACACCGGAGAUGGCACUCUAUAGAUGACUCUUUUAUCUGCUUUCCAUUCAUGUUGGCUUUUCACUGAGUCAGCGAUCAAGGUUAUUACAGGCAUUCCAUCCUGGCCAUCUUAUAGCC ----------........((((((((........((((((.(((..(((((.....)).....((((((((......))))))))))).))))))))).((((........))))))))))))...... ( -31.30, z-score = -1.84, R) >droSec1.super_0 4853072 119 - 21120651 ----------UUACACCGGAGAUGGCACUCUACAGAUGACUCUUUUAUCUGCUUUCCAUUCAUGUUGGCUUUUCACUGAGUCAGCGAUCAAGGUUAUUACAGGCAUUCCAUCCUGGCCAUCUUAUAGCC ----------........((((((((......(((((((.....)))))))....((.....(((((((((......))))))))).....))......((((........))))))))))))...... ( -32.90, z-score = -2.39, R) >droYak2.chr3L 12736119 119 - 24197627 ----------UUACUCCGGAGAUGGCACUCUACAGAUGACUCUUUUAUCUGCCUUCCAUUCAUGUUGGCUUUUCACUGAGUCAGCGAUCAAGGUUAUUACAGGCAUUCCAUCCUGGCCAUCUUAUAACC ----------........((((((((......(((((((.....)))))))((((....((..((((((((......))))))))))..))))......((((........))))))))))))...... ( -33.70, z-score = -2.84, R) >droEre2.scaffold_4784 12672386 119 - 25762168 ----------UUACACCGGAGAUGGCACUCUACAGAUGACUCUUUUAUCUGCUUGCCAUUCAUGUUGGCUUUUCCCUGAGUCAGCGAUCAAGGUUAUUACAGGCAUUCCAUCCUGGCCAUCUUAUAGCC ----------........((((((((......(((((((.....)))))))...(((.....(((((((((......))))))))).....))).....((((........))))))))))))...... ( -34.50, z-score = -2.22, R) >consensus __________UUACACCGGAGAUGGCACUCUACAGAUGACUCUUUUAUCUGCUUUCCAUUCAUGUUGGCUUUUCACUGAGUCAGCGAUCAAGGUUAUUACAGGCAUUCCAUCCUGGCCAUCUUAUAGCC ..................((((((((........(.((((((...................................)))))).).......(((......)))...........))))))))...... (-13.08 = -12.66 + -0.42)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:21:44 2011