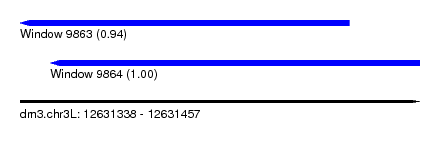
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 12,631,338 – 12,631,457 |
| Length | 119 |
| Max. P | 0.997095 |

| Location | 12,631,338 – 12,631,436 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 83.28 |
| Shannon entropy | 0.31197 |
| G+C content | 0.49078 |
| Mean single sequence MFE | -28.87 |
| Consensus MFE | -17.84 |
| Energy contribution | -19.17 |
| Covariance contribution | 1.34 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.45 |
| SVM RNA-class probability | 0.939354 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
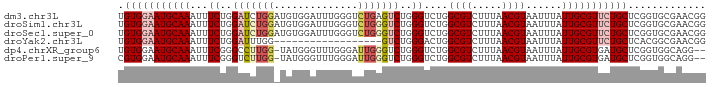
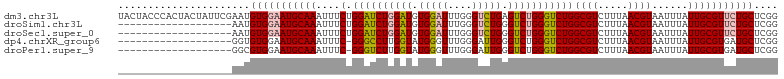
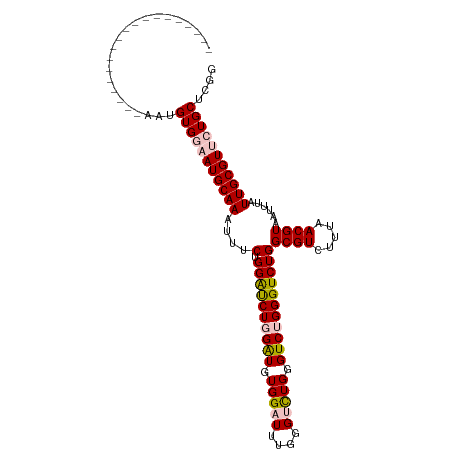
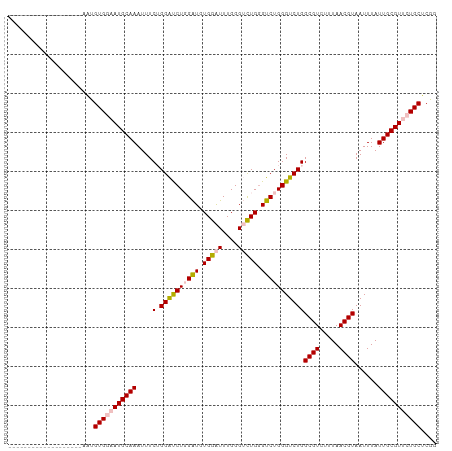
>dm3.chr3L 12631338 98 - 24543557 UGUGGAAUGCAAAUUUCUGGAUCUGGAUGUGGAUUUGGGUCUGAGUCUGGGUCUGGCGUCUUUAACGUAAUUUAUUGCGUUCUGCUCGGUGCGAACGG .(..((((((((......(((((..(((.(((((....))))).)))..))))).((((.....))))......))))))))..).((.......)). ( -31.40, z-score = -2.99, R) >droSim1.chr3L 12017059 98 - 22553184 UGUGGAAUGCAAAUUUCUGGAUCUGGAUGUGGAUUUGGGUCUGGGUCUGGGUCUGGCGUCUUUAACGUAAUUUAUUGCGUUCUGCUCGGUGCGAACGG .(..((((((((......(((((..(((.(((((....))))).)))..))))).((((.....))))......))))))))..).((.......)). ( -33.50, z-score = -3.78, R) >droSec1.super_0 4817624 98 - 21120651 UGUGGAAUGCAAAUUUCUGGAUCUGGAUGUGGAUUUGGGUCUGGGUCUGGGUCUGGCGUCUUUAACGUAAUUUAUUGCGUUCUGCUCGGUGCGAACGG .(..((((((((......(((((..(((.(((((....))))).)))..))))).((((.....))))......))))))))..).((.......)). ( -33.50, z-score = -3.78, R) >droYak2.chr3L 12698119 80 - 24197627 UGUGGAAUGCAAAUUUCUGGAUUUGG------------------GUCUGGGACUGGCGUCUUUAACGUAAUUUAUUGCGUUCUGCUCACGGCGAACGG .(..((((((((..(((..(((....------------------)))..)))...((((.....))))......))))))))..)...((.....)). ( -23.40, z-score = -2.25, R) >dp4.chrXR_group6 3415421 95 - 13314419 UGUGGAAUGCAAAUUUCGGGCCUUGG-UAUGGGUUUGGGAUUGGGUCUGGGUCUGGCGUCUUUAACGUAAUUUAUUGCGUGAUGCUCGGUGGCAGG-- ........((.(((..(((((((...-...)))))))..)))..))(((...((((((((....((((((....))))))))))).)))...))).-- ( -27.80, z-score = -1.50, R) >droPer1.super_9 1714373 95 - 3637205 CGUGGAAUGCAAAUUUCGGGUCUUGG-UAUGGGUUUGGGAUUGGGUCUGGGUCUGGCGUCUUUAACGUAAUUUAUUGCGUGAUGCUCGGUGGCAGG-- ........((.(((..(((..((...-...))..)))..)))..))(((...((((((((....((((((....))))))))))).)))...))).-- ( -23.60, z-score = -0.65, R) >consensus UGUGGAAUGCAAAUUUCUGGAUCUGG_UGUGGAUUUGGGUCUGGGUCUGGGUCUGGCGUCUUUAACGUAAUUUAUUGCGUUCUGCUCGGUGCGAACGG .(((((((((((...((..(((((((..............)))))))..))....((((.....))))......)))))))))))............. (-17.84 = -19.17 + 1.34)
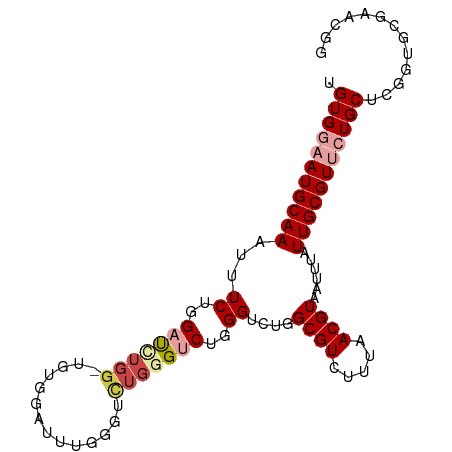
| Location | 12,631,347 – 12,631,457 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 83.76 |
| Shannon entropy | 0.25031 |
| G+C content | 0.47371 |
| Mean single sequence MFE | -29.84 |
| Consensus MFE | -24.00 |
| Energy contribution | -24.72 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.13 |
| Mean z-score | -3.32 |
| Structure conservation index | 0.80 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.04 |
| SVM RNA-class probability | 0.997095 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 12631347 110 - 24543557 UACUACCCACUACUAUUCGAAUGUGGAAUGCAAAUUUCUGGAUCUGGAUGUGGAUUUGGGUCUGAGUCUGGGUCUGGCGUCUUUAACGUAAUUUAUUGCGUUCUGCUCGG .................(((..(..((((((((......(((((..(((.(((((....))))).)))..))))).((((.....))))......))))))))..)))). ( -32.80, z-score = -2.68, R) >droSim1.chr3L 12017068 91 - 22553184 -------------------AAUGUGGAAUGCAAAUUUCUGGAUCUGGAUGUGGAUUUGGGUCUGGGUCUGGGUCUGGCGUCUUUAACGUAAUUUAUUGCGUUCUGCUCGG -------------------...(..((((((((......(((((..(((.(((((....))))).)))..))))).((((.....))))......))))))))..).... ( -33.20, z-score = -5.38, R) >droSec1.super_0 4817633 91 - 21120651 -------------------AAUGUGGAAUGCAAAUUUCUGGAUCUGGAUGUGGAUUUGGGUCUGGGUCUGGGUCUGGCGUCUUUAACGUAAUUUAUUGCGUUCUGCUCGG -------------------...(..((((((((......(((((..(((.(((((....))))).)))..))))).((((.....))))......))))))))..).... ( -33.20, z-score = -5.38, R) >dp4.chrXR_group6 3415428 90 - 13314419 -------------------GGUGUGGAAUGCAAAUUUC-GGGCCUUGGUAUGGGUUUGGGAUUGGGUCUGGGUCUGGCGUCUUUAACGUAAUUUAUUGCGUGAUGCUCGG -------------------((..((((.....(((..(-((((((......)))))))..)))...))))..)).((((((....((((((....))))))))))))... ( -25.00, z-score = -1.41, R) >droPer1.super_9 1714380 90 - 3637205 -------------------GGCGUGGAAUGCAAAUUUC-GGGUCUUGGUAUGGGUUUGGGAUUGGGUCUGGGUCUGGCGUCUUUAACGUAAUUUAUUGCGUGAUGCUCGG -------------------(((.((((.....(((..(-((..((......))..)))..)))...)))).))).((((((....((((((....))))))))))))... ( -25.00, z-score = -1.74, R) >consensus ___________________AAUGUGGAAUGCAAAUUUCUGGAUCUGGAUGUGGAUUUGGGUCUGGGUCUGGGUCUGGCGUCUUUAACGUAAUUUAUUGCGUUCUGCUCGG ......................(((((((((((......((((((((((.(((((....))))).)))))))))).((((.....))))......))))))))))).... (-24.00 = -24.72 + 0.72)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:21:42 2011