
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 12,628,114 – 12,628,212 |
| Length | 98 |
| Max. P | 0.982608 |

| Location | 12,628,114 – 12,628,212 |
|---|---|
| Length | 98 |
| Sequences | 7 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 83.41 |
| Shannon entropy | 0.33018 |
| G+C content | 0.36671 |
| Mean single sequence MFE | -23.86 |
| Consensus MFE | -15.07 |
| Energy contribution | -16.64 |
| Covariance contribution | 1.57 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.75 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.11 |
| SVM RNA-class probability | 0.982608 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
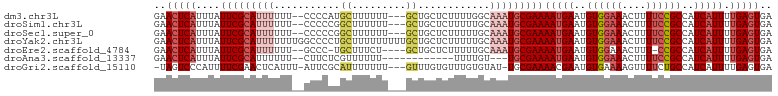
>dm3.chr3L 12628114 98 + 24543557 GAACUCAUUUAUUCGCAUUUUUU--CCCCAUGCUUUUUU---GCUGCUCUUUUGGCAAAUGCGAAAAUGAAUGUGGAAACUUUUCCGCCAUCAUUUUGAGUGA ..(((((....(((((((((...--......((......---)).(((.....))))))))))))(((((..((((((....))))))..))))).))))).. ( -26.70, z-score = -3.31, R) >droSim1.chr3L 12013619 98 + 22553184 GAACUCAUUUAUUCGCAUUUUUU--CCCCCGGCUUUUUU---GCUGCUCUUUUUGCAAAUGCGAAAAUGAAUGUGGAAACUUUUCCGCCAUCAUUUUGAGUGA ..(((((....(((((((((...--.....(((......---)))((.......)))))))))))(((((..((((((....))))))..))))).))))).. ( -27.70, z-score = -3.81, R) >droSec1.super_0 4814230 98 + 21120651 GAACUCAUUUAUUCGCAUUUUUU--CCCCCGGCUUUUUU---GCUGCUCUUUUUGCAAAUGCGAAAAUGAAUGUGGAAACUUUUCCGCCAUCAUUUUGAGUGA ..(((((....(((((((((...--.....(((......---)))((.......)))))))))))(((((..((((((....))))))..))))).))))).. ( -27.70, z-score = -3.81, R) >droYak2.chr3L 12694114 103 + 24197627 GAACUCAUUUAUUCGCAUUUUUUUGGCCCCUGCUUUUUUUUUGCUGCUCUUUUUGCAAAUGCGAAAAUGAAUGUGGAAACUUUUCCGCCAUCAUUUUGAGUGA ..(((((....(((((((((....(((....)))...........((.......)))))))))))(((((..((((((....))))))..))))).))))).. ( -25.60, z-score = -2.81, R) >droEre2.scaffold_4784 12632586 95 + 25762168 GAACUCAUUUAUUCGCAUUUUUU--GCCC-UGCUUUCU----GCUGCUCUUUUUGCAAAUGCGAAAAUGAAUGUGGAAACUUU-CCGCCAUCAUUUUGAGUGA ..(((((....(((((((((...--....-.((.....----)).((.......)))))))))))(((((..(((((.....)-))))..))))).))))).. ( -25.20, z-score = -3.21, R) >droAna3.scaffold_13337 19896049 86 - 23293914 GAACUCAUUUAUUCGCAUUUUUU--CUUCUCGUUUUUU------------UUUUGU---UGCGAAAAUGAAUGUGGAAACUUUUCCGCCAUCAUUUUGAGUGA ..(((((....((((((......--.............------------......---))))))(((((..((((((....))))))..))))).))))).. ( -19.31, z-score = -2.55, R) >droGri2.scaffold_15110 7681540 97 - 24565398 -UAGUCCCAUUUUCGAACUCAUUU-AUUCGCAUUUUUUU---GUUUGUGUUUGUGUAU-UGCGAAAACGAAUGUGAAAAGUUUUCUGCCAUCAUUUUGAGUGA -...............(((((...-.(((((((((((((---((..(((......)))-.))))))..)))))))))..((.....))........))))).. ( -14.80, z-score = 0.29, R) >consensus GAACUCAUUUAUUCGCAUUUUUU__CCCCCUGCUUUUUU___GCUGCUCUUUUUGCAAAUGCGAAAAUGAAUGUGGAAACUUUUCCGCCAUCAUUUUGAGUGA ..(((((....(((((((((...........((.........))............)))))))))(((((..((((((....))))))..))))).))))).. (-15.07 = -16.64 + 1.57)

| Location | 12,628,114 – 12,628,212 |
|---|---|
| Length | 98 |
| Sequences | 7 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 83.41 |
| Shannon entropy | 0.33018 |
| G+C content | 0.36671 |
| Mean single sequence MFE | -20.57 |
| Consensus MFE | -13.74 |
| Energy contribution | -14.82 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.632134 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
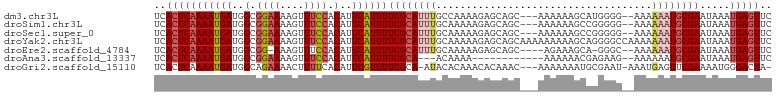
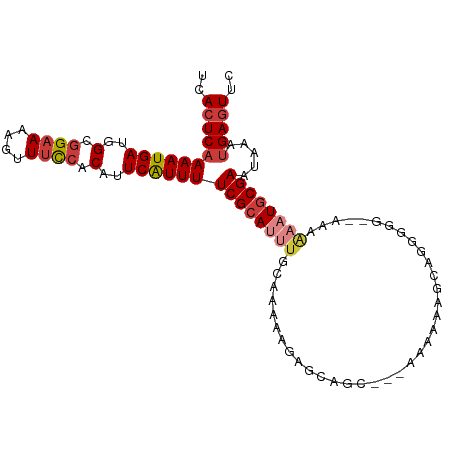
>dm3.chr3L 12628114 98 - 24543557 UCACUCAAAAUGAUGGCGGAAAAGUUUCCACAUUCAUUUUCGCAUUUGCCAAAAGAGCAGC---AAAAAAGCAUGGGG--AAAAAAUGCGAAUAAAUGAGUUC ..(((((((((((((..((((....)))).)).))))))((((((((.((......((...---......))....))--...)))))))).....))))).. ( -24.00, z-score = -2.03, R) >droSim1.chr3L 12013619 98 - 22553184 UCACUCAAAAUGAUGGCGGAAAAGUUUCCACAUUCAUUUUCGCAUUUGCAAAAAGAGCAGC---AAAAAAGCCGGGGG--AAAAAAUGCGAAUAAAUGAGUUC ..(((((((((((((..((((....)))).)).))))))((((((((((.......)).((---......))......--...)))))))).....))))).. ( -22.90, z-score = -1.61, R) >droSec1.super_0 4814230 98 - 21120651 UCACUCAAAAUGAUGGCGGAAAAGUUUCCACAUUCAUUUUCGCAUUUGCAAAAAGAGCAGC---AAAAAAGCCGGGGG--AAAAAAUGCGAAUAAAUGAGUUC ..(((((((((((((..((((....)))).)).))))))((((((((((.......)).((---......))......--...)))))))).....))))).. ( -22.90, z-score = -1.61, R) >droYak2.chr3L 12694114 103 - 24197627 UCACUCAAAAUGAUGGCGGAAAAGUUUCCACAUUCAUUUUCGCAUUUGCAAAAAGAGCAGCAAAAAAAAAGCAGGGGCCAAAAAAAUGCGAAUAAAUGAGUUC ..(((((((((((((..((((....)))).)).))))))((((((((((.......)).((.........))...........)))))))).....))))).. ( -22.80, z-score = -1.77, R) >droEre2.scaffold_4784 12632586 95 - 25762168 UCACUCAAAAUGAUGGCGG-AAAGUUUCCACAUUCAUUUUCGCAUUUGCAAAAAGAGCAGC----AGAAAGCA-GGGC--AAAAAAUGCGAAUAAAUGAGUUC ..(((((((((((((..((-(.....))).)).))))))((((((((((.......)).((----.....)).-....--...)))))))).....))))).. ( -22.70, z-score = -1.80, R) >droAna3.scaffold_13337 19896049 86 + 23293914 UCACUCAAAAUGAUGGCGGAAAAGUUUCCACAUUCAUUUUCGCA---ACAAAA------------AAAAAACGAGAAG--AAAAAAUGCGAAUAAAUGAGUUC ..(((((((((((((..((((....)))).)).))))))(((((---......------------.............--......))))).....))))).. ( -15.71, z-score = -1.89, R) >droGri2.scaffold_15110 7681540 97 + 24565398 UCACUCAAAAUGAUGGCAGAAAACUUUUCACAUUCGUUUUCGCA-AUACACAAACACAAAC---AAAAAAAUGCGAAU-AAAUGAGUUCGAAAAUGGGACUA- ...((((...((....))......(((((..(((((((((((((-................---.......)))))..-))))))))..)))))))))....- ( -13.00, z-score = -0.55, R) >consensus UCACUCAAAAUGAUGGCGGAAAAGUUUCCACAUUCAUUUUCGCAUUUGCAAAAAGAGCAGC___AAAAAAGCAGGGGG__AAAAAAUGCGAAUAAAUGAGUUC ..(((((((((((..(.((((....)))).)..))))))((((((((....................................)))))))).....))))).. (-13.74 = -14.82 + 1.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:21:40 2011