| Sequence ID | dm3.chr3L |
|---|---|
| Location | 12,613,097 – 12,613,208 |
| Length | 111 |
| Max. P | 0.706129 |

| Location | 12,613,097 – 12,613,208 |
|---|---|
| Length | 111 |
| Sequences | 10 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 70.13 |
| Shannon entropy | 0.62980 |
| G+C content | 0.61946 |
| Mean single sequence MFE | -31.97 |
| Consensus MFE | -16.30 |
| Energy contribution | -16.03 |
| Covariance contribution | -0.27 |
| Combinations/Pair | 1.47 |
| Mean z-score | -0.65 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.551777 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
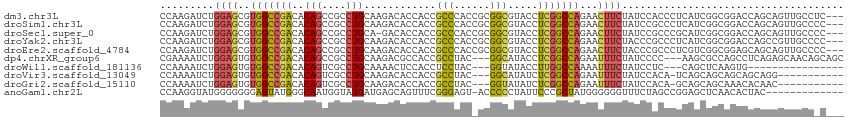
>dm3.chr3L 12613097 111 + 24543557 CCAAGAUCUGGAGCGUGGCCGACACAGCCGCCUGCAAGACACCACCGCCCACCGCGGCGUACCUCGGCCAGAACUUCUAUCCACCCUCAUCGGCGGACCAGCAGUUGCCUC--- (((.....))).((.(((((((....(((((..((...........)).....))))).....))))))).((((.((.(((.((......)).)))..)).))))))...--- ( -33.40, z-score = -0.14, R) >droSim1.chr3L 11998757 111 + 22553184 CCAAGAUCUGGAGCGUGGCCGACACAGCCGCCUGCAAGACACCACCGCCCACCGCGGCGUACCUCGGCCAGAACUUCUAUCCGCCCUCAUCGGCGGACCAGCAGUUGCCCC--- .........((.((.(((((((....(((((..((...........)).....))))).....))))))).((((.((.((((((......))))))..)).)))))))).--- ( -40.00, z-score = -1.82, R) >droSec1.super_0 4799331 110 + 21120651 CCAAGAUCUGGAGCGUGGCCGACACAGCCGCCUGCA-GACACCACCGCCCACCGCGGCGUACCUCGGCCAGAACUUCUAUCCGCCCGCAUCGGCGGACCAGCAGUUGCCCC--- .........((.((.(((((((....(((((..((.-(......).)).....))))).....))))))).((((.((.((((((......))))))..)).)))))))).--- ( -40.40, z-score = -1.56, R) >droYak2.chr3L 12678001 111 + 24197627 CCAAGAUCUGGAGCGUGGCCGACACAGCCGCCUGCAAGACACCACCGCCCACCGCGGCGUACCUCGGCCAGAACUUCUACCCGCCCUCAUCGGCGGACCAGCCGUUGCCCC--- .(((...((((....(((((((....(((((..((...........)).....))))).....)))))))..........(((((......))))).))))...)))....--- ( -36.80, z-score = -0.80, R) >droEre2.scaffold_4784 12617453 111 + 25762168 CCAAGAUCUGGAGCGUGGCCGACACAGCCGCCUGCAAGACACCACCGCCCACCGCGGCGUACCUCGGCCAGAACUUCUACCCGCCCUCGUCGGCGGAGCAGCAGUUGCCCC--- .........((.((.(((((((....(((((..((...........)).....))))).....))))))).((((.((.((((((......))))).).)).)))))))).--- ( -39.20, z-score = -0.64, R) >dp4.chrXR_group6 3396889 108 + 13314419 CGAAAAUCUGGAGUGUGGCCGACACAGCCGCCUGCAAGACGCCACCGCCUAC---GGCAUACCUCGGCCAGAAUUUCUAUCCCC---AAGCGCCAGCCUCAGAGCAACAGCAGC ......(((((..(.(((((((....((((...((...........))...)---))).....))))))).)............---..((....)).)))))((....))... ( -26.30, z-score = 0.89, R) >droWil1.scaffold_181136 2285941 92 - 2313701 CCAAAAUCUGGAGUGUGGCCGACACAGUCGCCUGCAAAACUCCACCUCCUAC---GGUAUACCUUGGCCAAAAUUUCUAUCCUC---CAGCUCAAGUG---------------- .......((((((..(((((((..(((....))).........(((......---))).....)))))))...........)))---)))........---------------- ( -19.92, z-score = -1.17, R) >droVir3.scaffold_13049 6018363 99 - 25233164 CCAAAAUCUGGAGUGUGGCCGACACAGUCGCCUGCAAGACACCACCGCCUAC---GGCAUAUCUCGGCCAGAAUUUCUAUCCACA-UCAGCAGCAGCAGCAGG----------- ((.....(((..(((.((((((....(((........)))......(((...---))).....))))))((.....))...))).-.)))..((....)).))----------- ( -24.80, z-score = -0.08, R) >droGri2.scaffold_15110 7665387 99 - 24565398 CCAAAAUCUGGAGUGUGGCCGACACAGUCGCCUGCAAGACACCACCGCCUAC---GGUAUAUCUCGGCCAGAAUUUCUAUCCACA-GCAGCAGCAAACACAAC----------- (((.....))).((((((((((....(((........)))...((((....)---))).....))))))................-((....))..))))...----------- ( -25.90, z-score = -1.67, R) >anoGam1.chr2L 7502200 100 + 48795086 CCAAGGUAUGGGGGGGAGUAUGGGCAAUGGUAUGAUGAGCAGUUUCGGGAGU-ACCCCCUAUUCCCGCUAUGGGGGGUUUCUAGCCGGAGCUCAACACUAC------------- ..........((.(((((((.(((....(((((..((((....))))...))-))))))))))))).)).((..(((((((.....)))))))..))....------------- ( -33.00, z-score = 0.48, R) >consensus CCAAGAUCUGGAGCGUGGCCGACACAGCCGCCUGCAAGACACCACCGCCCAC_GCGGCGUACCUCGGCCAGAACUUCUAUCCACCCUCAGCGGCAGACCAGCAGUUGCC_C___ .........((((..(((((((..(((....)))............(((......))).....)))))))...))))..................................... (-16.30 = -16.03 + -0.27)


| Location | 12,613,097 – 12,613,208 |
|---|---|
| Length | 111 |
| Sequences | 10 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 70.13 |
| Shannon entropy | 0.62980 |
| G+C content | 0.61946 |
| Mean single sequence MFE | -42.50 |
| Consensus MFE | -22.99 |
| Energy contribution | -22.61 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.58 |
| Mean z-score | -0.69 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.706129 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
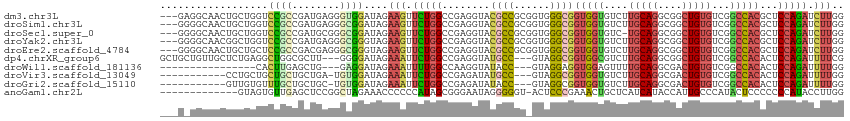
>dm3.chr3L 12613097 111 - 24543557 ---GAGGCAACUGCUGGUCCGCCGAUGAGGGUGGAUAGAAGUUCUGGCCGAGGUACGCCGCGGUGGGCGGUGGUGUCUUGCAGGCGGCUGUGUCGGCCACGCUCCAGAUCUUGG ---(..((((((.(((.((((((......))))))))).)))).(((((((..((.(((((.(..((((....)).))..)..)))))))..))))))).))..)......... ( -46.90, z-score = -0.16, R) >droSim1.chr3L 11998757 111 - 22553184 ---GGGGCAACUGCUGGUCCGCCGAUGAGGGCGGAUAGAAGUUCUGGCCGAGGUACGCCGCGGUGGGCGGUGGUGUCUUGCAGGCGGCUGUGUCGGCCACGCUCCAGAUCUUGG ---(((((((((.(((.((((((......))))))))).)))).(((((((..((.(((((.(..((((....)).))..)..)))))))..))))))).)))))......... ( -54.10, z-score = -1.58, R) >droSec1.super_0 4799331 110 - 21120651 ---GGGGCAACUGCUGGUCCGCCGAUGCGGGCGGAUAGAAGUUCUGGCCGAGGUACGCCGCGGUGGGCGGUGGUGUC-UGCAGGCGGCUGUGUCGGCCACGCUCCAGAUCUUGG ---(((((((((.(((.((((((......))))))))).)))).(((((((..((.(((((.(..((((....))))-..)..)))))))..))))))).)))))......... ( -56.90, z-score = -2.02, R) >droYak2.chr3L 12678001 111 - 24197627 ---GGGGCAACGGCUGGUCCGCCGAUGAGGGCGGGUAGAAGUUCUGGCCGAGGUACGCCGCGGUGGGCGGUGGUGUCUUGCAGGCGGCUGUGUCGGCCACGCUCCAGAUCUUGG ---(((((...((((.(((((((......)))))))...)))).(((((((..((.(((((.(..((((....)).))..)..)))))))..))))))).)))))......... ( -50.30, z-score = -0.18, R) >droEre2.scaffold_4784 12617453 111 - 25762168 ---GGGGCAACUGCUGCUCCGCCGACGAGGGCGGGUAGAAGUUCUGGCCGAGGUACGCCGCGGUGGGCGGUGGUGUCUUGCAGGCGGCUGUGUCGGCCACGCUCCAGAUCUUGG ---(((((((((.((((.(((((......))))))))).)))).(((((((..((.(((((.(..((((....)).))..)..)))))))..))))))).)))))......... ( -56.10, z-score = -1.46, R) >dp4.chrXR_group6 3396889 108 - 13314419 GCUGCUGUUGCUCUGAGGCUGGCGCUU---GGGGAUAGAAAUUCUGGCCGAGGUAUGCC---GUAGGCGGUGGCGUCUUGCAGGCGGCUGUGUCGGCCACACUCCAGAUUUUCG ((.(((...(((....))).)))))((---((((..((.....))((((((..((.(((---((..((((.......))))..)))))))..))))))...))))))....... ( -38.50, z-score = 1.01, R) >droWil1.scaffold_181136 2285941 92 + 2313701 ----------------CACUUGAGCUG---GAGGAUAGAAAUUUUGGCCAAGGUAUACC---GUAGGAGGUGGAGUUUUGCAGGCGACUGUGUCGGCCACACUCCAGAUUUUGG ----------------..((((.((..---(((........)))..)))))).......---...((((((((.....(((((....)))))....)))).))))......... ( -30.20, z-score = -1.76, R) >droVir3.scaffold_13049 6018363 99 + 25233164 -----------CCUGCUGCUGCUGCUGA-UGUGGAUAGAAAUUCUGGCCGAGAUAUGCC---GUAGGCGGUGGUGUCUUGCAGGCGACUGUGUCGGCCACACUCCAGAUUUUGG -----------...((....))..(((.-.(((((.......))(((((((((((((((---(....)))).)))))..((((....)))).))))))))))..)))....... ( -33.10, z-score = -0.17, R) >droGri2.scaffold_15110 7665387 99 + 24565398 -----------GUUGUGUUUGCUGCUGC-UGUGGAUAGAAAUUCUGGCCGAGAUAUACC---GUAGGCGGUGGUGUCUUGCAGGCGACUGUGUCGGCCACACUCCAGAUUUUGG -----------.((.((((..(......-.)..)))).))....(((((((((((((((---(....)))).)))))..((((....)))).)))))))............... ( -33.10, z-score = -0.74, R) >anoGam1.chr2L 7502200 100 - 48795086 -------------GUAGUGUUGAGCUCCGGCUAGAAACCCCCCAUAGCGGGAAUAGGGGGU-ACUCCCGAAACUGCUCAUCAUACCAUUGCCCAUACUCCCCCCCAUACCUUGG -------------((((((.((((((((.((((...........)))).)))...((((..-..))))......)))))......))))))............(((.....))) ( -25.80, z-score = 0.16, R) >consensus ___G_GGCAACUGCUGGUCCGCCGAUGAGGGCGGAUAGAAGUUCUGGCCGAGGUACGCCGC_GUGGGCGGUGGUGUCUUGCAGGCGGCUGUGUCGGCCACGCUCCAGAUCUUGG ..................((((........))))....(((.(((((........((((......))))(((((....(((((....)))))...)))))...))))).))).. (-22.99 = -22.61 + -0.38)
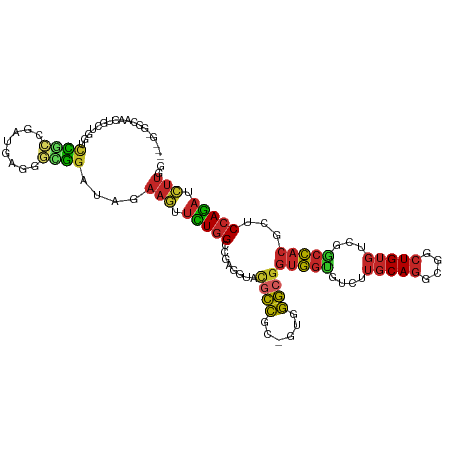

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:21:35 2011