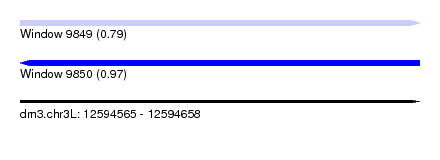
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 12,594,565 – 12,594,658 |
| Length | 93 |
| Max. P | 0.972739 |

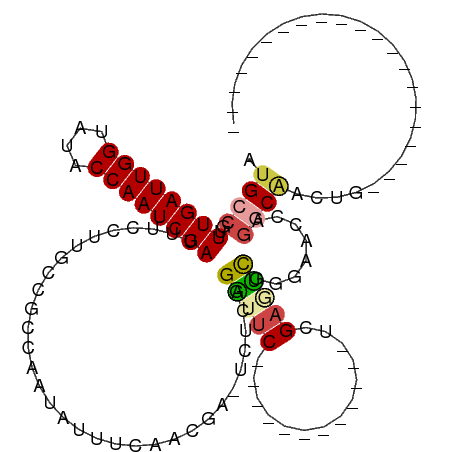
| Location | 12,594,565 – 12,594,658 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 70.51 |
| Shannon entropy | 0.50209 |
| G+C content | 0.49105 |
| Mean single sequence MFE | -28.17 |
| Consensus MFE | -12.36 |
| Energy contribution | -12.84 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.785958 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 12594565 93 + 24543557 AUGCCGUUUGAUUGGUAUACCAAUCGAUUUCCUUGCCGCCAAUAUUUCAAUGA-UCUCGGAUUC------------UCGAGUCUGGAACCAGGCAACUGGCAACUG-------------- .((((..((((((((....)))))))).....(((((.......(((((..((-.(((((....------------))))))))))))...)))))..))))....-------------- ( -27.90, z-score = -2.58, R) >droSim1.chr3L 11980062 86 + 22553184 AUGCCGUUUGAUUGGUAUACCAAUCGAUUUCCUUGCCGCCAAUAUUUCAACGA-UCUCGGAUUC------------UCGAGUCGGGAACCAGGCAACUG--------------------- .((((..((((((((....)))))))).(((((((....)))........(((-.(((((....------------))))))))))))...))))....--------------------- ( -25.80, z-score = -2.23, R) >droSec1.super_0 4780876 80 + 21120651 AUGCCGUUUGAUUGGUAUACCAAUCGAUUUCCUUGCCGCCAAUAUUUCAACGA-UCUCGGAUUC------------UGGAGUC------CAGGCAACUG--------------------- .((((..((((((((....))))))))..........................-....(((((.------------...))))------).))))....--------------------- ( -21.10, z-score = -1.66, R) >droEre2.scaffold_4784 12598482 119 + 25762168 AUGCCGUUUGAUUGGUAUACCAAUCGAUUUCCUUGCCGCCAAUACUUCAACGA-UCUCGGAUUCCGAAGAGGGCCCUUGAAUCUGGAACCAGGCAACUGGCCACUGCCAACUGGCAACUG .((((..((((((((....)))))))).....(((((................-((((((...))).)))((..((........))..)).))))).((((....))))...)))).... ( -36.10, z-score = -2.01, R) >droAna3.scaffold_13337 19864848 86 - 23293914 AUGCCGUUUGAUUGGUAUACCAAUCGAUUUCCUUGCCGCCAAUAUUUCAACGG-UCUCGGGUUCGG--------UCUCGAUUCGGUUUCGAACCG------------------------- ..((((((.((((((....))))))((.....(((....)))....)))))))-)....(((((((--------..(((...)))..))))))).------------------------- ( -24.30, z-score = -1.17, R) >dp4.chrXR_group6 3378968 103 + 13314419 AUGCCGUUUGAUUGGUAUACCAAUCGAUUUACUUGCCGCCAAUAUUUCAACGAGUCCCCGGGACUGAGACUGGAACUGGGACUGAGACCGGGACCGGGGACCG----------------- ..((.((((((((((....)))))))).......)).))...........((.((((((((..(((.(.((((........)).)).))))..))))))))))----------------- ( -33.81, z-score = -1.53, R) >consensus AUGCCGUUUGAUUGGUAUACCAAUCGAUUUCCUUGCCGCCAAUAUUUCAACGA_UCUCGGAUUC____________UCGAGUCGGGAACCAGGCAACUG_____________________ .((((..((((((((....))))))))................................(((((..............)))))........))))......................... (-12.36 = -12.84 + 0.48)

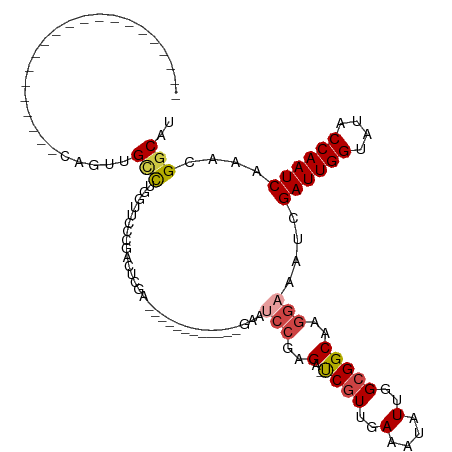
| Location | 12,594,565 – 12,594,658 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 70.51 |
| Shannon entropy | 0.50209 |
| G+C content | 0.49105 |
| Mean single sequence MFE | -30.12 |
| Consensus MFE | -16.54 |
| Energy contribution | -17.15 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.87 |
| SVM RNA-class probability | 0.972739 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 12594565 93 - 24543557 --------------CAGUUGCCAGUUGCCUGGUUCCAGACUCGA------------GAAUCCGAGA-UCAUUGAAAUAUUGGCGGCAAGGAAAUCGAUUGGUAUACCAAUCAAACGGCAU --------------....((((.((((((..(((.((((((((.------------.....)))).-))..)).)))...)))))).........((((((....))))))....)))). ( -30.20, z-score = -2.77, R) >droSim1.chr3L 11980062 86 - 22553184 ---------------------CAGUUGCCUGGUUCCCGACUCGA------------GAAUCCGAGA-UCGUUGAAAUAUUGGCGGCAAGGAAAUCGAUUGGUAUACCAAUCAAACGGCAU ---------------------....((((.((((.((..((((.------------.....)))).-((((..(....)..))))...)).))))((((((....))))))....)))). ( -28.00, z-score = -2.31, R) >droSec1.super_0 4780876 80 - 21120651 ---------------------CAGUUGCCUG------GACUCCA------------GAAUCCGAGA-UCGUUGAAAUAUUGGCGGCAAGGAAAUCGAUUGGUAUACCAAUCAAACGGCAU ---------------------....((((..------.......------------...(((..(.-((((..(....)..)))))..)))....((((((....))))))....)))). ( -22.80, z-score = -1.68, R) >droEre2.scaffold_4784 12598482 119 - 25762168 CAGUUGCCAGUUGGCAGUGGCCAGUUGCCUGGUUCCAGAUUCAAGGGCCCUCUUCGGAAUCCGAGA-UCGUUGAAGUAUUGGCGGCAAGGAAAUCGAUUGGUAUACCAAUCAAACGGCAU ((.(((((....)))))))(((.((((((((.(((..((((...(((...((....)).)))..))-))...))).))..)))))).........((((((....))))))....))).. ( -38.90, z-score = -1.05, R) >droAna3.scaffold_13337 19864848 86 + 23293914 -------------------------CGGUUCGAAACCGAAUCGAGA--------CCGAACCCGAGA-CCGUUGAAAUAUUGGCGGCAAGGAAAUCGAUUGGUAUACCAAUCAAACGGCAU -------------------------(((((((....))))))).(.--------(((...((..(.-((((..(....)..)))))..)).....((((((....))))))...)))).. ( -30.10, z-score = -3.30, R) >dp4.chrXR_group6 3378968 103 - 13314419 -----------------CGGUCCCCGGUCCCGGUCUCAGUCCCAGUUCCAGUCUCAGUCCCGGGGACUCGUUGAAAUAUUGGCGGCAAGUAAAUCGAUUGGUAUACCAAUCAAACGGCAU -----------------.(((((((((..(.((.((.............)).).).)..)))))))))(((..(....)..)))....((.....((((((....)))))).....)).. ( -30.72, z-score = -0.87, R) >consensus _____________________CAGUUGCCUGGUUCCCGACUCGA____________GAAUCCGAGA_UCGUUGAAAUAUUGGCGGCAAGGAAAUCGAUUGGUAUACCAAUCAAACGGCAU ..........................(((..............................(((.....((((..(....)..))))...)))....((((((....))))))....))).. (-16.54 = -17.15 + 0.61)

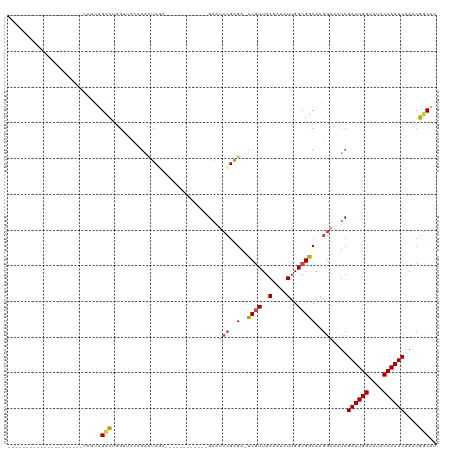
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:21:30 2011