| Sequence ID | dm3.chr2L |
|---|---|
| Location | 5,063,917 – 5,064,082 |
| Length | 165 |
| Max. P | 0.906658 |

| Location | 5,063,917 – 5,064,082 |
|---|---|
| Length | 165 |
| Sequences | 6 |
| Columns | 187 |
| Reading direction | reverse |
| Mean pairwise identity | 71.21 |
| Shannon entropy | 0.50733 |
| G+C content | 0.47814 |
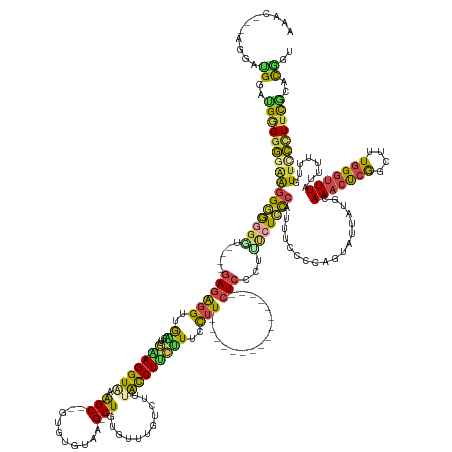
| Mean single sequence MFE | -58.55 |
| Consensus MFE | -23.84 |
| Energy contribution | -22.99 |
| Covariance contribution | -0.85 |
| Combinations/Pair | 1.78 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.906658 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
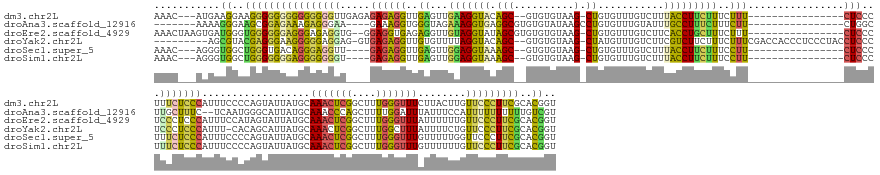
>dm3.chr2L 5063917 165 - 23011544 AAAC---AUGAAGGAAGGGGGGGGGGGGGGUUGAGAGAGAGGUUGAGUUGAAGGUACAGC--GUGUGUAAG-CUGUGUUUGUCUUUACCUUCUUUCUUU----------------CUCCCUUUCUCCCAUUUCCCCAGUAUUAUGCAAACUCGGCUUUGGGUUUCUUACUUGUUCCCUUCGCACGGU ....---..(((((..(((((..(((((((..((((((.((((.(((....(..((((((--........)-)))))..)..))).))))))))))..)----------------))))))..))))).....(((((......((.......)).)))))..............)))))....... ( -63.40, z-score = -2.54, R) >droAna3.scaffold_12916 13701186 158 - 16180835 -------AAAAGGGAAGCGGAGAAAGAGGGAA----GAAAGGUGGGUAGAAAGGUGAGGCGUGUGUAUAAGCCUGUGUUUGUAUUUGCCUUUCUUUCUU----------------CUGGCUUGCUUUC--UCAAUGGGCAUUAUGCAAACCCAGCUUUUGGAUUUAUUUCCAUUUUUUUUUUGUCGU -------((((((((((((.((..((((((((----((((((..((((.(((..(.((((..........)))))..))).))))..))))))))))))----------------))..)))))))))--....((((...........)))).)))))(((......)))................ ( -53.40, z-score = -2.96, R) >droEre2.scaffold_4929 5143676 168 - 26641161 AAACUAAGUGAUGGGUGGGGGGAGGGAGAGGUG--GGAGGUGAGAGGUUGUAGGUAUAGCGUGUGUGUAAG-CUGUGUUUGUCUUCACCUGCUUUCUUU----------------CUCCCUCCCUCCCAUUUCCAUAGUAUUAUGCAAACUCGGCUUUGGGUUUAUUUUUUGUUCCCUUCGCACGGU ..((((...((((((.((((((.((((((((.(--(.((((((.(((...((..((((((..........)-)))))..))))))))))).)).)))))----------------))))))))).))))))....))))...........(((((...(((..((.....))..)))...)).))). ( -64.10, z-score = -3.18, R) >droYak2.chr2L 11863279 173 + 22324452 ---------AGCGUACGAGGGAAGGGGGAGGAG-GUGAGAGGUUGUGUUUUAGGUACAGC--GUGUGUAAG-CUAUGUUUGUCUUCGUCUUCUUUCUUUCGACCACCCUCCCUACCUCCCUCCCUCCCAUUU-CACAGCAUUAUGCAAACUCGGCUUUGGCUUUAUUUUCUGUUCCCUUCGCACGGU ---------..(((.((((((((((((((((((-(((.((((.((.(((..(((...(((--(.(..((((-(...)))))..).)).)).....)))..))))).))))..))))).))))))))......-...(((.....((.......))....)))..........))))).))).))).. ( -51.40, z-score = -0.25, R) >droSec1.super_5 3150978 161 - 5866729 AAAC---AGGGUGGCUGGGUGACAGGGAGGUU----GAGAGGUUGAGUUGGAGGUAAAGC--GUGUGUAAG-CUGUGUUUGUCUUUACCUUCUUUCCUU----------------CUCCCUUUCUCCCAUUUCCCCAGUAUUAUGCAAACUCGGCUUUGGGUUUGUUUUUGGUUCCCUUCGCACGGU ....---((((..((((((.((..((((((..----(.((((..(((..(((((((((((--(.(..(...-..)..).)).)))))))))).)))..)----------------))).)..))))))...))))))))((((.(((((((((....)))))))))...)))).))))......... ( -55.60, z-score = -1.86, R) >droSim1.chr2L 4899572 161 - 22036055 AAAC---AGGGUGGCUGGGGGGGAGGGGGGGU----GAGAGGUUGAGUUGGAGGUAAAGC--GUGUGUAAG-CUGUGUUUGUCUUUACCUUCUUUCCUU----------------CUCCCUUUCUCCCAUUUCCCCAGUAUUAUGCAAACUCGGCUUUGGGUUUGUUUUUUGUUCCCUUCGCACGGU ....---((((..((((((((((((..((((.----((.(((.......(((((((((((--(.(..(...-..)..).)).))))))))))...))))----------------).))))..)))))....))))))).....(((((((((....)))))))))........))))......... ( -63.40, z-score = -2.67, R) >consensus AAAC___AGGAUGGAUGGGGGGAAGGGGGGGU____GAGAGGUUGAGUUGAAGGUAAAGC__GUGUGUAAG_CUGUGUUUGUCUUUACCUUCUUUCCUU________________CUCCCUUUCUCCCAUUUCCCCAGUAUUAUGCAAACUCGGCUUUGGGUUUAUUUUUUGUUCCCUUCGCACGGU ...........((..(((((((((.(((((........((((((((((((((((((.((...(..((......))..)....)).))))))).............................................((.....)).)))))))))))........))))).)))))))))..)).. (-23.84 = -22.99 + -0.85)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:17:53 2011