
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 12,558,151 – 12,558,241 |
| Length | 90 |
| Max. P | 0.803470 |

| Location | 12,558,151 – 12,558,241 |
|---|---|
| Length | 90 |
| Sequences | 7 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 71.28 |
| Shannon entropy | 0.55935 |
| G+C content | 0.48533 |
| Mean single sequence MFE | -17.77 |
| Consensus MFE | -7.70 |
| Energy contribution | -7.90 |
| Covariance contribution | 0.21 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.803470 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 12558151 90 - 24543557 -------CCUCACCCCCUUCAAGUCGUAGACAAAGUGGCAGAAACAUGCAUAAUUAAUUUAUGCCCCUCUGGGCGCACCUC--CCGCUCCUCUUCUCCA -------...............(..(.(((...(((((.((.....(((((((.....))))((((....))))))).)).--)))))..))).)..). ( -16.10, z-score = -0.73, R) >droEre2.scaffold_4784 12560841 97 - 25762168 CCACCCCUACCUUCCCCGCCAAGUCGUAGACAAAGUGGCAGAAACAUGCAUAAUUAAUUUAUGCCCCUCCGGGCGCACCUC--CCGCUCUUCUGCUCCA ......(((((((.......)))..)))).....(..((((((....((((((.....))))))......(((((......--.)))))))))))..). ( -22.40, z-score = -2.31, R) >droYak2.chr3L 12621231 88 - 24197627 ---------CCGCCCCCGCCAAGUCGUAGACAAAGUGGCAGAAACAUGCAUAAUUAAUUUAUGCCCCUCCGGGCGCACCUU--CUGCUCUUCUCCUCCA ---------..((((..((((.(((...)))....))))........((((((.....))))))......))))(((....--.)))............ ( -22.00, z-score = -2.05, R) >droSec1.super_0 4744592 91 - 21120651 --------UCGUCACCUUCCAAGUCGUAGACAAAGUGGCAGAAACAUGCAUAAUUAAUUUAUGCCCCUCUGGGCGCACCUCCUCCGCUCCUCUUCUCCA --------..(((((..........)).)))..(((((.((.....(((((((.....))))((((....)))))))....)))))))........... ( -16.90, z-score = -0.85, R) >droSim1.chr3L 11941638 91 - 22553184 --------UCGUCCCCUUCGAAGUCGUAGACAAAGUGGCAGAAACAUGCAUAAUUAAUUUAUGCCCCUCUGGGCGCACCUCCUCCGCUCCUCUUCUCCA --------..(((.....((....))..)))..(((((.((.....(((((((.....))))((((....)))))))....)))))))........... ( -17.90, z-score = -0.81, R) >droWil1.scaffold_181136 2198615 69 + 2313701 -----------------UUACCCUCGUAGACAAAGAAGCAGAAACAUGUAUAAUUAAUUUAUGCUUUUUUGUUUUCCUUUUUUUUG------------- -----------------........(.(((((((((((((.....................))))))))))))).)..........------------- ( -12.20, z-score = -2.92, R) >droAna3.scaffold_13337 19826611 82 + 23293914 -----------CCGCACUCCGCCUCGUAGACAAAGUGGCAGAAACAUGCAUAAUUAAUUUAUGCCCCAUU---CUCACAGAGUCCUCUCUGCUUUC--- -----------.........(((.(.........).)))((((....((((((.....))))))....))---))..(((((....))))).....--- ( -16.90, z-score = -1.97, R) >consensus _________CCUCCCCCUCCAAGUCGUAGACAAAGUGGCAGAAACAUGCAUAAUUAAUUUAUGCCCCUCUGGGCGCACCUC_UCCGCUCCUCUUCUCCA ..................................(((((........((((((.....))))))........)).)))..................... ( -7.70 = -7.90 + 0.21)
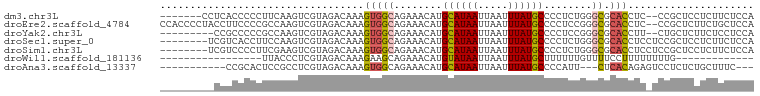
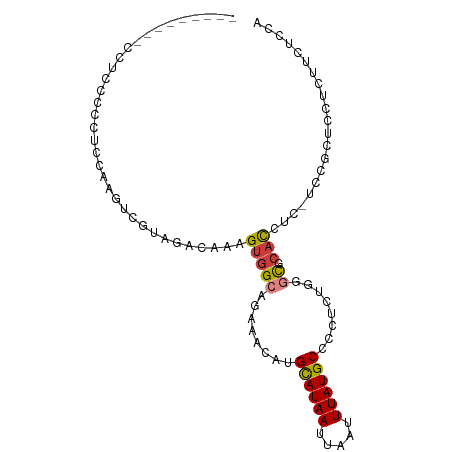

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:21:24 2011