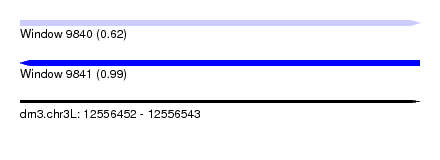
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 12,556,452 – 12,556,543 |
| Length | 91 |
| Max. P | 0.985228 |

| Location | 12,556,452 – 12,556,543 |
|---|---|
| Length | 91 |
| Sequences | 9 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 64.59 |
| Shannon entropy | 0.67531 |
| G+C content | 0.44335 |
| Mean single sequence MFE | -22.82 |
| Consensus MFE | -9.36 |
| Energy contribution | -9.60 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.03 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.619064 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
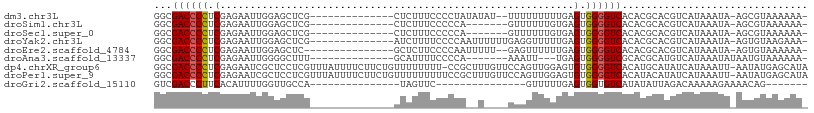
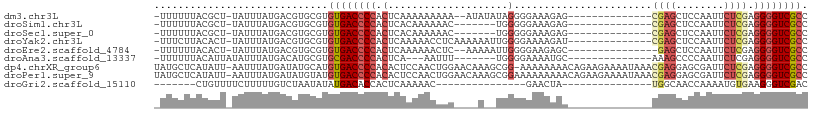
>dm3.chr3L 12556452 91 + 24543557 GGCGACCCCUCGAGAAUUGGAGCUCG--------------CUCUUUCCCCUAUAUAU--UUUUUUUUUGAGUGGGGUCACACGCACGUCAUAAAUA-AGCGUAAAAAA- .(.(((((((((((((..((((....--------------)))).............--....)))))))..)))))).)((((............-.))))......- ( -21.63, z-score = -0.84, R) >droSim1.chr3L 11939904 86 + 22553184 GGCGACCCCUCGAGAAUUGGAGCUCG--------------CUCUUUCCCCCA-------GUUUUUUGUGAGUGGGGUCACACGCACGUCAUAAAUA-AGCGUAAAAAA- .(.((((((.((((((((((......--------------.........)))-------)))))).....).)))))).)((((............-.))))......- ( -21.68, z-score = -0.24, R) >droSec1.super_0 4742859 86 + 21120651 GGCGACCCCUCGAGAAUUGGAGCUCG--------------CUCUUUCCCCCA-------GUUUUUUGUGAGUGGGGUCACACGCACGUCAUAAAUA-AGCGUAAAAAA- .(.((((((.((((((((((......--------------.........)))-------)))))).....).)))))).)((((............-.))))......- ( -21.68, z-score = -0.24, R) >droYak2.chr3L 12619542 93 + 24197627 GGCGACCCCUCGAGAAUUGGAGCUCG--------------AUCUUUUCCCCAAUUUUUUGAGGUUUUUGAGUGGGGUCACACGCACGUCAUAAAUA-AGUGUAAGAAA- .(.(((((((((((........))))--------------)((....((.(((....))).)).....))..)))))).)..((((..........-.))))......- ( -24.30, z-score = -0.61, R) >droEre2.scaffold_4784 12559255 90 + 25762168 GGCGACCCCUCGAGAAUUGGAGCUC---------------GCUCUUCCCCAAUUUUU--GAGUUUUUUGAGUGGGGUCACACGCACGUCAUAAAUA-AGUGUAAAAAA- .(.(((((((((((((((((.....---------------........)))))))))--))...........)))))).)..((((..........-.))))......- ( -24.02, z-score = -1.12, R) >droAna3.scaffold_13337 19825136 84 - 23293914 GGCGACCCCUCGAGAAUUGGGGCUUU--------------GCAUUUUCCCCA-------AAAUU---UGAGUGGGGUCGCACGCAUGUCAUAAAUAUAAUGUAAAAAA- .(((((((((((((..((((((....--------------.......)))))-------)..))---)))..))))))))..((((............))))......- ( -29.30, z-score = -3.07, R) >dp4.chrXR_group6 3339095 107 + 13314419 GGCGACCCCUCGAGAAUCGCUCCUCGUUUAUUUUCUUCUGUUUUUUUU-CCGCUUUGUUCCAGUUGGAGUGUGGGGUCACAUGCAUAUCAUAAAUU-AAUAUGAGCAUA .(.((((((.((((........))))......................-.((((..(.......)..)))).)))))).)((((...(((((....-..))))))))). ( -25.60, z-score = -1.34, R) >droPer1.super_9 1636074 108 + 3637205 GGCGACCCCUCGAGAAUCGCUCCUCGUUUAUUUUCUUCUGUUUUUUUUUCCGCUUUGUUCCAGUUGGAGUGUGGGGUCACAUACAUAUCAUAAAUU-AAUAUGAGCAUA .((((((((.((((........))))........................((((..(.......)..)))).)))))).........(((((....-..)))))))... ( -25.10, z-score = -1.48, R) >droGri2.scaffold_15110 7604346 72 - 24565398 GUCGACCCUUCACAUUUUGGUUGCCA---------------UAGUUC---------------GUUUUUGAGUGGUGUCAUAUAUUAGACAAAAAGAAAACAG------- (.(((((...........))))).).---------------......---------------((((((...(..((((........))))..).))))))..------- ( -12.10, z-score = -0.30, R) >consensus GGCGACCCCUCGAGAAUUGGAGCUCG______________CUCUUUCCCCCA__U____GAAUUUUGUGAGUGGGGUCACACGCACGUCAUAAAUA_AGUGUAAAAAA_ ...((((((.((((........))).....................................((....))).))))))............................... ( -9.36 = -9.60 + 0.24)
| Location | 12,556,452 – 12,556,543 |
|---|---|
| Length | 91 |
| Sequences | 9 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 64.59 |
| Shannon entropy | 0.67531 |
| G+C content | 0.44335 |
| Mean single sequence MFE | -24.51 |
| Consensus MFE | -10.90 |
| Energy contribution | -11.64 |
| Covariance contribution | 0.74 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.19 |
| SVM RNA-class probability | 0.985228 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
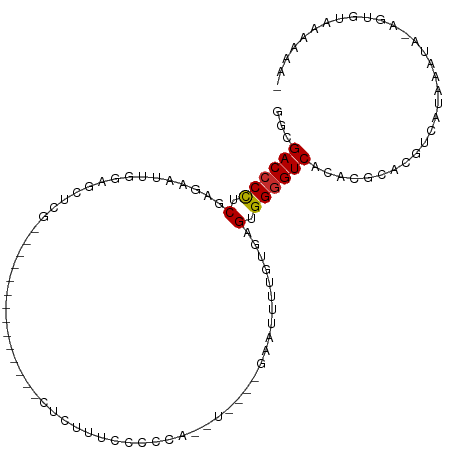
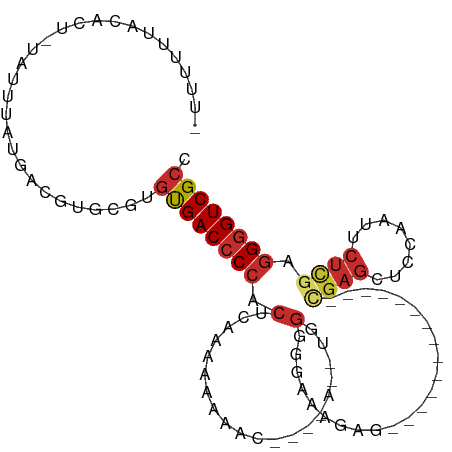
>dm3.chr3L 12556452 91 - 24543557 -UUUUUUACGCU-UAUUUAUGACGUGCGUGUGACCCCACUCAAAAAAAAA--AUAUAUAGGGGAAAGAG--------------CGAGCUCCAAUUCUCGAGGGGUCGCC -......((((.-............))))((((((((.((..........--......))(((((.(((--------------....)))...)))))..)))))))). ( -23.51, z-score = -1.18, R) >droSim1.chr3L 11939904 86 - 22553184 -UUUUUUACGCU-UAUUUAUGACGUGCGUGUGACCCCACUCACAAAAAAC-------UGGGGGAAAGAG--------------CGAGCUCCAAUUCUCGAGGGGUCGCC -......((((.-............))))((((((((.....((......-------)).(((((.(((--------------....)))...)))))..)))))))). ( -25.62, z-score = -1.20, R) >droSec1.super_0 4742859 86 - 21120651 -UUUUUUACGCU-UAUUUAUGACGUGCGUGUGACCCCACUCACAAAAAAC-------UGGGGGAAAGAG--------------CGAGCUCCAAUUCUCGAGGGGUCGCC -......((((.-............))))((((((((.....((......-------)).(((((.(((--------------....)))...)))))..)))))))). ( -25.62, z-score = -1.20, R) >droYak2.chr3L 12619542 93 - 24197627 -UUUCUUACACU-UAUUUAUGACGUGCGUGUGACCCCACUCAAAAACCUCAAAAAAUUGGGGAAAAGAU--------------CGAGCUCCAAUUCUCGAGGGGUCGCC -.......((((-((....))).)))...((((((((.........((((((....))))))......(--------------((((........))))))))))))). ( -28.00, z-score = -2.73, R) >droEre2.scaffold_4784 12559255 90 - 25762168 -UUUUUUACACU-UAUUUAUGACGUGCGUGUGACCCCACUCAAAAAACUC--AAAAAUUGGGGAAGAGC---------------GAGCUCCAAUUCUCGAGGGGUCGCC -.......((((-((....))).)))...((((((((.(((......(((--(.....))))...)))(---------------(((........)))).)))))))). ( -23.10, z-score = -1.02, R) >droAna3.scaffold_13337 19825136 84 + 23293914 -UUUUUUACAUUAUAUUUAUGACAUGCGUGCGACCCCACUCA---AAUUU-------UGGGGAAAAUGC--------------AAAGCCCCAAUUCUCGAGGGGUCGCC -............................((((((((..((.---....(-------(((((.......--------------....)))))).....)))))))))). ( -25.30, z-score = -2.50, R) >dp4.chrXR_group6 3339095 107 - 13314419 UAUGCUCAUAUU-AAUUUAUGAUAUGCAUGUGACCCCACACUCCAACUGGAACAAAGCGG-AAAAAAAACAGAAGAAAAUAAACGAGGAGCGAUUCUCGAGGGGUCGCC .(((((((((..-....)))))...))))((((((((....(((....))).........-......................((((((....)))))).)))))))). ( -31.50, z-score = -4.12, R) >droPer1.super_9 1636074 108 - 3637205 UAUGCUCAUAUU-AAUUUAUGAUAUGUAUGUGACCCCACACUCCAACUGGAACAAAGCGGAAAAAAAAACAGAAGAAAAUAAACGAGGAGCGAUUCUCGAGGGGUCGCC .(((((((((..-....)))))...))))((((((((....(((....)))................................((((((....)))))).)))))))). ( -29.50, z-score = -3.50, R) >droGri2.scaffold_15110 7604346 72 + 24565398 -------CUGUUUUCUUUUUGUCUAAUAUAUGACACCACUCAAAAAC---------------GAACUA---------------UGGCAACCAAAAUGUGAAGGGUCGAC -------............((((........))))...........(---------------((.((.---------------(.(((.......))).).)).))).. ( -8.40, z-score = 0.86, R) >consensus _UUUUUUACACU_UAUUUAUGACGUGCGUGUGACCCCACUCAAAAAAAAC____A__UGGGGGAAAGAG______________CGAGCUCCAAUUCUCGAGGGGUCGCC .............................((((((((.(....................).......................((((........)))).)))))))). (-10.90 = -11.64 + 0.74)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:21:23 2011