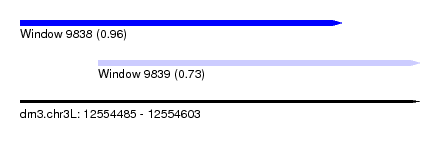
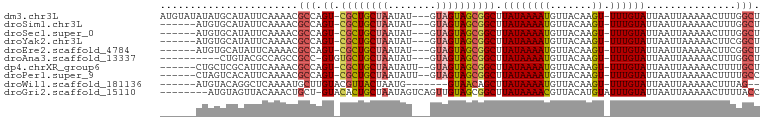
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 12,554,485 – 12,554,603 |
| Length | 118 |
| Max. P | 0.956611 |

| Location | 12,554,485 – 12,554,580 |
|---|---|
| Length | 95 |
| Sequences | 10 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 67.80 |
| Shannon entropy | 0.69289 |
| G+C content | 0.39106 |
| Mean single sequence MFE | -24.42 |
| Consensus MFE | -10.19 |
| Energy contribution | -9.63 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.70 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.63 |
| SVM RNA-class probability | 0.956611 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
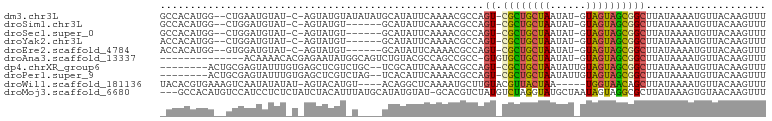
>dm3.chr3L 12554485 95 + 24543557 GCCACAUGG--CUGAAUGUAU-C-AGUAUGUAUAUAUGCAUAUUCAAAACGCCAGU-CGCUGCUAAUAU-GUAGUAGCGGCUUAUAAAAUGUUACAAGUUU (((....))--)....((((.-(-((((((((....)))))))..........(((-((((((((....-.))))))))))).......)).))))..... ( -26.90, z-score = -2.03, R) >droSim1.chr3L 11937955 89 + 22553184 GCCACAUGG--CUGGAUGUAU-C-AGUAUGU------GCAUAUUCAAAACGCCAGU-CGCUGCUAAUAU-GUAGUAGCGGCUUAUAAAAUGUUACAAGUUU (((....((--((((.(((..-.-(((((..------..)))))....))))))))-)(((((((....-.)))))))))).................... ( -25.80, z-score = -1.66, R) >droSec1.super_0 4740927 89 + 21120651 GCCACAUGG--CUGGAUGUAU-C-AGUAUGU------GCAUAUUCAAAACGCCAGU-CGCUGCUAAUAU-GUAGUAGCGGCUUAUAAAAUGUUACAAGUUU (((....((--((((.(((..-.-(((((..------..)))))....))))))))-)(((((((....-.)))))))))).................... ( -25.80, z-score = -1.66, R) >droYak2.chr3L 12617527 89 + 24197627 ACCACAUGG--CUGGAUGUAU-C-AGUAUGU------GCAUAUUCAAAACGCCAGU-CGCUGCUAAUAU-GUAGUAGCGGCUUAUAAAAUGUUACAAGUUU ...((((((--((((((((((-.-.....))------)))..))))....)))(((-((((((((....-.)))))))))))......))))......... ( -23.70, z-score = -1.16, R) >droEre2.scaffold_4784 12557438 89 + 25762168 ACCACAUGG--GUGGAUGUAU-C-AGUAUGU------GCAUAUUCAAAACGCCAGU-CGCUGCUAAUAU-GUAGUAGCGGCUUAUAAAAUGUUACAAGUUU ...((((.(--(((.((((((-.-.....))------))))........))))(((-((((((((....-.)))))))))))......))))......... ( -24.40, z-score = -1.47, R) >droAna3.scaffold_13337 19823304 85 - 23293914 --------------ACAAAACACGAGAAUAUGGCAGUCUGUACGCCAGCCGCC-GUGUGCUGCUAAUAU-GUAGUAGCGGCUUAUAAAAUGUUACAAGUUU --------------....((((.....(((((((.(.(((.....)))).)))-))))(((((((....-....)))))))........))))........ ( -22.20, z-score = -0.50, R) >dp4.chrXR_group6 3336699 90 + 13314419 --------ACUGCGAGUAUUUGUGAGCUCGUCUGC--UCGCAUUCAAAACGCCAGU-CGCUGCUAAUAUUGUAGUAGCGGCUUAUAAAAUGUUACAAGUUU --------...(((......(((((((......))--))))).......))).(((-((((((((......)))))))))))................... ( -29.52, z-score = -2.87, R) >droPer1.super_9 1633666 90 + 3637205 --------ACUGCGAGUAUUUGUGAGCUCGUCUAG--UCACAUUCAAAACGCCAGU-CGCUGCUAAUAUUGUAGUAGCGGCUUAUAAAAUGUUACAAGUUU --------...((((((........))))))...(--(.(((((.........(((-((((((((......))))))))))).....))))).))...... ( -24.84, z-score = -1.96, R) >droWil1.scaffold_181136 2191604 91 - 2313701 UACACGUGAAAGUCAAUAUAUAU-AGUACAUGU----ACAGGCUCAAAAUGCUUGUACGUUACUAA-----UGGUAACAGCUUAUAAAAUGUUACAAGUUU ......................(-((((.((((----((((((.......))))))))))))))).-----..((((((..........))))))...... ( -22.40, z-score = -2.36, R) >droMoj3.scaffold_6680 7013618 97 + 24764193 ---GCCACAUGUCCAUCCUCUCUAUCUACAUUUAUGCAUAUGUAU-GCACGUCUAUGUCUAGGUAUGCUAAUAGUAGGCGCUUAUAAAGUGUAACAAGUUU ---...((.(((.............((((..(((.((((((.((.-(((......))).)).)))))))))..))))(((((.....))))).))).)).. ( -18.60, z-score = -0.08, R) >consensus _CCACAUGG__CUGAAUGUAU_C_AGUAUGU_U____GCAUAUUCAAAACGCCAGU_CGCUGCUAAUAU_GUAGUAGCGGCUUAUAAAAUGUUACAAGUUU ..................................................(((.....(((((((......)))))))))).................... (-10.19 = -9.63 + -0.56)
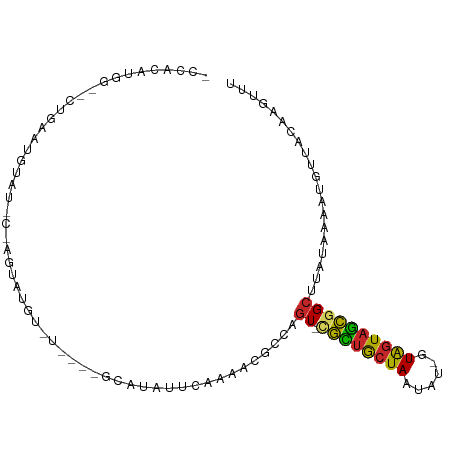
| Location | 12,554,508 – 12,554,603 |
|---|---|
| Length | 95 |
| Sequences | 10 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 83.90 |
| Shannon entropy | 0.33331 |
| G+C content | 0.32786 |
| Mean single sequence MFE | -22.46 |
| Consensus MFE | -11.63 |
| Energy contribution | -12.26 |
| Covariance contribution | 0.63 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.732479 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
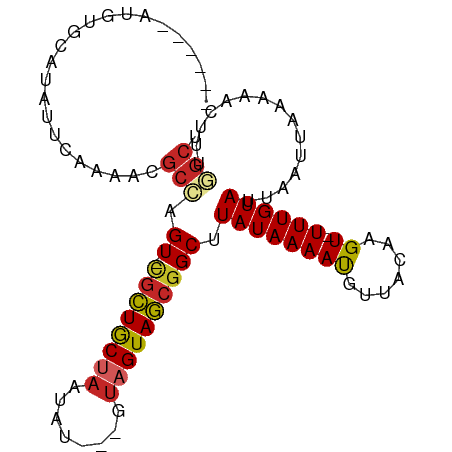
>dm3.chr3L 12554508 95 + 24543557 AUGUAUAUAUGCAUAUUCAAAACGCCAGU-CGCUGCUAAUAU---GUAGUAGCGGCUUAUAAAAUGUUACAAGU-UUUGUAUUAAUUAAAAACUUUGGCU (((((....))))).........((((((-((((((((....---.)))))))))).((((((((.......))-))))))..............)))). ( -25.70, z-score = -2.90, R) >droSim1.chr3L 11937978 89 + 22553184 ------AUGUGCAUAUUCAAAACGCCAGU-CGCUGCUAAUAU---GUAGUAGCGGCUUAUAAAAUGUUACAAGU-UUUGUAUUAAUUAAAAACUUUGGCU ------.................((((((-((((((((....---.)))))))))).((((((((.......))-))))))..............)))). ( -24.10, z-score = -2.63, R) >droSec1.super_0 4740950 89 + 21120651 ------AUGUGCAUAUUCAAAACGCCAGU-CGCUGCUAAUAU---GUAGUAGCGGCUUAUAAAAUGUUACAAGU-UUUGUAUUAAUUAAAAACUUUGGCU ------.................((((((-((((((((....---.)))))))))).((((((((.......))-))))))..............)))). ( -24.10, z-score = -2.63, R) >droYak2.chr3L 12617550 89 + 24197627 ------AUGUGCAUAUUCAAAACGCCAGU-CGCUGCUAAUAU---GUAGUAGCGGCUUAUAAAAUGUUACAAGU-UUUGUAUUAAUUAAAAACUUCGGCU ------.................((((((-((((((((....---.)))))))))))((((((((.......))-))))))...............))). ( -24.10, z-score = -2.83, R) >droEre2.scaffold_4784 12557461 89 + 25762168 ------AUGUGCAUAUUCAAAACGCCAGU-CGCUGCUAAUAU---GUAGUAGCGGCUUAUAAAAUGUUACAAGU-UUUGUAUUAAUUAAAAACUUCGGCU ------.................((((((-((((((((....---.)))))))))))((((((((.......))-))))))...............))). ( -24.10, z-score = -2.83, R) >droAna3.scaffold_13337 19823327 85 - 23293914 ----------CUGUACGCCAGCCGCC-GUGUGCUGCUAAUAU---GUAGUAGCGGCUUAUAAAAUGUUACAAGU-UUUGUAUUAAUUAAAAACUUUGGCU ----------......(((((..(((-((.((((((......---))))))))))).((((((((.......))-)))))).............))))). ( -25.10, z-score = -2.22, R) >dp4.chrXR_group6 3336722 90 + 13314419 ------CUGCUCGCAUUCAAAACGCCAGU-CGCUGCUAAUAUU--GUAGUAGCGGCUUAUAAAAUGUUACAAGU-UUUGUAUUAAUUAAAAACUUUUGCU ------......(((...........(((-((((((((.....--.)))))))))))((((((((.......))-))))))...............))). ( -21.40, z-score = -1.92, R) >droPer1.super_9 1633689 90 + 3637205 ------CUAGUCACAUUCAAAACGCCAGU-CGCUGCUAAUAUU--GUAGUAGCGGCUUAUAAAAUGUUACAAGU-UUUGUAUUAAUUAAAAACUUUUGCC ------...((.(((((.........(((-((((((((.....--.))))))))))).....))))).))((((-(((..........)))))))..... ( -20.94, z-score = -2.55, R) >droWil1.scaffold_181136 2191632 84 - 2313701 ------AUGUACAGGCUCAAAAUGCUUGUACGUUACUAAUG-------GUAACAGCUUAUAAAAUGUUACAAGU-UUUGUAUUAAUUAAAAACUUUAG-- ------((((((((((.......))))))))))........-------((((((..........))))))((((-(((..........)))))))...-- ( -20.10, z-score = -2.82, R) >droGri2.scaffold_15110 16962791 91 + 24565398 --------AUGUAGUUACAAACUGCU-GUACACUGCUAAUAGUCAGUUGUAGCGGCUUAUAAAACGUUACAUGUAUUUGUAUUAAUUAAAAACUUUUACC --------..(((((((((......)-))).)))))...(((((((.(((((((..........))))))).....))).))))................ ( -15.00, z-score = 0.54, R) >consensus ______AUGUGCAUAUUCAAAACGCCAGU_CGCUGCUAAUAU___GUAGUAGCGGCUUAUAAAAUGUUACAAGU_UUUGUAUUAAUUAAAAACUUUGGCU .......................(((.((.((((((((........))))))))))..............((((.((((.......)))).)))).))). (-11.63 = -12.26 + 0.63)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:21:22 2011