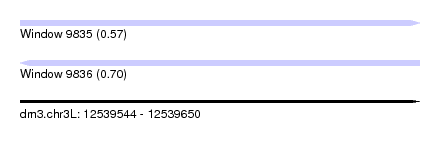
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 12,539,544 – 12,539,650 |
| Length | 106 |
| Max. P | 0.697413 |

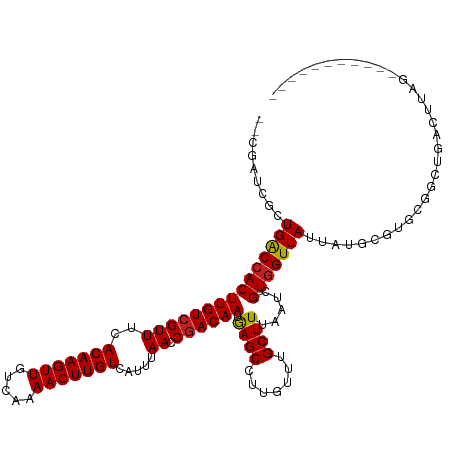
| Location | 12,539,544 – 12,539,650 |
|---|---|
| Length | 106 |
| Sequences | 12 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 86.87 |
| Shannon entropy | 0.28205 |
| G+C content | 0.43902 |
| Mean single sequence MFE | -28.64 |
| Consensus MFE | -21.43 |
| Energy contribution | -21.27 |
| Covariance contribution | -0.15 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.568146 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 12539544 106 + 24543557 --CGAUCGCUGGCCACUUGUCGUUUCACAAGUUGUCAAAACUUGUCAUUUAACCGACAAUGAGCCUUGUUUGCUUUAAUCGUGGUUAUUAUGCGUGCGGCUGACUUAG----------- --((..(((.(((((.((((((((..(((((((.....))))))).....)).)))))))).)))......((..((((....))))....))..)))..))......----------- ( -26.10, z-score = -0.67, R) >droSim1.chr3L 11922275 106 + 22553184 --CGAUCGCUGGCCACUUGUCGUUUCACAAGUUGUCAAAACUUGUCAUUUAACCGACAAUGAGCCUUGUUUGCUUUAAUCGUGGUUAUUAUGCGUGCGGCUGACUUAG----------- --((..(((.(((((.((((((((..(((((((.....))))))).....)).)))))))).)))......((..((((....))))....))..)))..))......----------- ( -26.10, z-score = -0.67, R) >droSec1.super_0 4726046 106 + 21120651 --CGAUCGCUGGCCACUUGUCGUUUCACAAGUUGUCAAAACUUGUCAUUUAACCGACAAUGAGCCUUGUUUGCUUUAAUCGUGGUUAUUAUGCGUGCGGCUGACUUAG----------- --((..(((.(((((.((((((((..(((((((.....))))))).....)).)))))))).)))......((..((((....))))....))..)))..))......----------- ( -26.10, z-score = -0.67, R) >droEre2.scaffold_4784 12541347 106 + 25762168 --CGAUCGCUGGCCACUUGUCGUUUCACAAGUUGUCAAAACUUGUCAUUUAACCGACAAUGAGCCUUGUUUGCUUUAAUCGUGGUUAUUAUGCGUGCGGCUGACUUAG----------- --((..(((.(((((.((((((((..(((((((.....))))))).....)).)))))))).)))......((..((((....))))....))..)))..))......----------- ( -26.10, z-score = -0.67, R) >droYak2.chr3L 12601867 106 + 24197627 --CGAUCGCUGGCCACUUGUCGUUUCACAAGUUGUCAAAACUUGUCAUUUAACCGACAAUGAGCCUUGUUUGCUUUAAUCGUGGUUAUUAUGCGUGCGGCUGACUUAG----------- --((..(((.(((((.((((((((..(((((((.....))))))).....)).)))))))).)))......((..((((....))))....))..)))..))......----------- ( -26.10, z-score = -0.67, R) >droAna3.scaffold_13337 19809887 106 - 23293914 --CGAUCGCUGACCACUUGUCGUUUCACAAGUUGUCAAAACUUGUCAUUUAACCGACAAUGAGCCUUGUUUGCUUUAAUCGUGGUUAUUAUGCGUGCGGCUGACUUAG----------- --((..((((((((((((((((((..(((((((.....))))))).....)).)))))).((((.......)))).....)))))))....)))..))..........----------- ( -26.30, z-score = -1.12, R) >dp4.chrXR_group6 3321784 117 + 13314419 --CAGCCGCUGACCACUUGUCGUUUCACAAGUUGUCAAAACUUGUCAUUUAACCGACAAUGAGCCUCGUUGGCUUUAAUCGUGGUUAUUAUGCGUGCGGCUGACUUGGGUCUCUCUGCC --(((((((((((.((((((......)))))).))))......(.(((.((((((.....(((((.....)))))......))))))..))))..))))))).....(((......))) ( -39.70, z-score = -3.38, R) >droPer1.super_9 1618654 117 + 3637205 --CAGCCGCUGACCACUUGUCGUUUCACAAGUUGUCAAAACUUGUCAUUUAACCGACAAUGAGCCUCGUUGGCUUUAAUCGUGGUUAUUAUGCGUGCGGCUGACUUGGGUCUCUCUGCC --(((((((((((.((((((......)))))).))))......(.(((.((((((.....(((((.....)))))......))))))..))))..))))))).....(((......))) ( -39.70, z-score = -3.38, R) >droWil1.scaffold_181136 2153489 114 - 2313701 --CAGCCGCUGACCACUUGUCGUUUCACAAGUUGUCAAAACUUGUCAUUUAACCUACAAUAAGCCUUGUUUGCUUUAAUCGUAGUUAUUAUGCGUUCAGGAGGUUUGGAAUGAUCA--- --.......((((.((((((......)))))).))))......((((((((((((.........((((..(((..((((....))))....)))..)))))))))..)))))))..--- ( -24.10, z-score = -0.42, R) >droVir3.scaffold_13049 5938481 114 - 25233164 CAUUGGCAGUGACCACUUGUCGUUUCACAAGUUGUCAAAACUUGUCAUUUAACCGACAAUGAGCCUUGUUUGCUUUAAUCGUGGUUAUUAUGCGUUCGCCCGAAUAAGUUUGCC----- ....((((((((((((((((((((..(((((((.....))))))).....)).)))))).((((.......)))).....)))))))))....((((....))))......)))----- ( -29.70, z-score = -2.03, R) >droMoj3.scaffold_6680 17734253 104 - 24764193 CACUGGCAGUGACCACUUGUCGUUUCACAAGUUGUCAAAACUUGUCAUUUAACCGACAAUGAGCCUUGUUUGCUUUAAUCGUGGUUAUUAUGCGUUCGCCCGAA--------------- .....(((((((((((((((((((..(((((((.....))))))).....)).)))))).((((.......)))).....))))))))..)))...........--------------- ( -26.60, z-score = -1.56, R) >droGri2.scaffold_15110 7585043 109 - 24565398 ------CACUGACCACUUGUCGUUUCACAAGUUGUCAAAACUUGUCAUUUAACCGACAAUGAGCCUUGUUUGCUUUAAUCGUGGUUAUUAUGCAUUCGACUGAAUAAGUCUGCCA---- ------...(((((((((((((((..(((((((.....))))))).....)).)))))).((((.......)))).....)))))))....(((...((((.....)))))))..---- ( -27.10, z-score = -2.32, R) >consensus __CGAUCGCUGACCACUUGUCGUUUCACAAGUUGUCAAAACUUGUCAUUUAACCGACAAUGAGCCUUGUUUGCUUUAAUCGUGGUUAUUAUGCGUGCGGCUGACUUAG___________ .........(((((((((((((((..(((((((.....))))))).....)).)))))).((((.......)))).....)))))))................................ (-21.43 = -21.27 + -0.15)

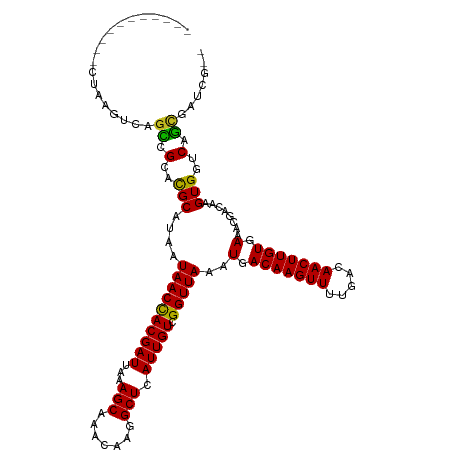
| Location | 12,539,544 – 12,539,650 |
|---|---|
| Length | 106 |
| Sequences | 12 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 86.87 |
| Shannon entropy | 0.28205 |
| G+C content | 0.43902 |
| Mean single sequence MFE | -28.57 |
| Consensus MFE | -19.88 |
| Energy contribution | -19.30 |
| Covariance contribution | -0.58 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.697413 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 12539544 106 - 24543557 -----------CUAAGUCAGCCGCACGCAUAAUAACCACGAUUAAAGCAAACAAGGCUCAUUGUCGGUUAAAUGACAAGUUUUGACAACUUGUGAAACGACAAGUGGCCAGCGAUCG-- -----------....((..(((((........((((((((((...(((.......))).))))).)))))..(.(((((((.....))))))).)........)))))..)).....-- ( -26.20, z-score = -1.47, R) >droSim1.chr3L 11922275 106 - 22553184 -----------CUAAGUCAGCCGCACGCAUAAUAACCACGAUUAAAGCAAACAAGGCUCAUUGUCGGUUAAAUGACAAGUUUUGACAACUUGUGAAACGACAAGUGGCCAGCGAUCG-- -----------....((..(((((........((((((((((...(((.......))).))))).)))))..(.(((((((.....))))))).)........)))))..)).....-- ( -26.20, z-score = -1.47, R) >droSec1.super_0 4726046 106 - 21120651 -----------CUAAGUCAGCCGCACGCAUAAUAACCACGAUUAAAGCAAACAAGGCUCAUUGUCGGUUAAAUGACAAGUUUUGACAACUUGUGAAACGACAAGUGGCCAGCGAUCG-- -----------....((..(((((........((((((((((...(((.......))).))))).)))))..(.(((((((.....))))))).)........)))))..)).....-- ( -26.20, z-score = -1.47, R) >droEre2.scaffold_4784 12541347 106 - 25762168 -----------CUAAGUCAGCCGCACGCAUAAUAACCACGAUUAAAGCAAACAAGGCUCAUUGUCGGUUAAAUGACAAGUUUUGACAACUUGUGAAACGACAAGUGGCCAGCGAUCG-- -----------....((..(((((........((((((((((...(((.......))).))))).)))))..(.(((((((.....))))))).)........)))))..)).....-- ( -26.20, z-score = -1.47, R) >droYak2.chr3L 12601867 106 - 24197627 -----------CUAAGUCAGCCGCACGCAUAAUAACCACGAUUAAAGCAAACAAGGCUCAUUGUCGGUUAAAUGACAAGUUUUGACAACUUGUGAAACGACAAGUGGCCAGCGAUCG-- -----------....((..(((((........((((((((((...(((.......))).))))).)))))..(.(((((((.....))))))).)........)))))..)).....-- ( -26.20, z-score = -1.47, R) >droAna3.scaffold_13337 19809887 106 + 23293914 -----------CUAAGUCAGCCGCACGCAUAAUAACCACGAUUAAAGCAAACAAGGCUCAUUGUCGGUUAAAUGACAAGUUUUGACAACUUGUGAAACGACAAGUGGUCAGCGAUCG-- -----------....((((((.....))....((((((((((...(((.......))).))))).)))))..)))).....(((((.((((((......)))))).)))))......-- ( -25.90, z-score = -1.39, R) >dp4.chrXR_group6 3321784 117 - 13314419 GGCAGAGAGACCCAAGUCAGCCGCACGCAUAAUAACCACGAUUAAAGCCAACGAGGCUCAUUGUCGGUUAAAUGACAAGUUUUGACAACUUGUGAAACGACAAGUGGUCAGCGGCUG-- ........(((....)))((((((..((((..((((((((((...((((.....)))).))))).))))).))).)......((((.((((((......)))))).)))))))))).-- ( -38.70, z-score = -3.35, R) >droPer1.super_9 1618654 117 - 3637205 GGCAGAGAGACCCAAGUCAGCCGCACGCAUAAUAACCACGAUUAAAGCCAACGAGGCUCAUUGUCGGUUAAAUGACAAGUUUUGACAACUUGUGAAACGACAAGUGGUCAGCGGCUG-- ........(((....)))((((((..((((..((((((((((...((((.....)))).))))).))))).))).)......((((.((((((......)))))).)))))))))).-- ( -38.70, z-score = -3.35, R) >droWil1.scaffold_181136 2153489 114 + 2313701 ---UGAUCAUUCCAAACCUCCUGAACGCAUAAUAACUACGAUUAAAGCAAACAAGGCUUAUUGUAGGUUAAAUGACAAGUUUUGACAACUUGUGAAACGACAAGUGGUCAGCGGCUG-- ---.............((..((((.(((.......(((((((..((((.......)))))))))))(((...(.(((((((.....))))))).)...)))..))).)))).))...-- ( -25.40, z-score = -1.26, R) >droVir3.scaffold_13049 5938481 114 + 25233164 -----GGCAAACUUAUUCGGGCGAACGCAUAAUAACCACGAUUAAAGCAAACAAGGCUCAUUGUCGGUUAAAUGACAAGUUUUGACAACUUGUGAAACGACAAGUGGUCACUGCCAAUG -----(((((((((..(((.((....))....((((((((((...(((.......))).))))).)))))..))).))))))((((.((((((......)))))).))))..))).... ( -30.10, z-score = -2.19, R) >droMoj3.scaffold_6680 17734253 104 + 24764193 ---------------UUCGGGCGAACGCAUAAUAACCACGAUUAAAGCAAACAAGGCUCAUUGUCGGUUAAAUGACAAGUUUUGACAACUUGUGAAACGACAAGUGGUCACUGCCAGUG ---------------....(((((((.(((..((((((((((...(((.......))).))))).))))).)))....))))((((.((((((......)))))).))))..))).... ( -27.50, z-score = -1.60, R) >droGri2.scaffold_15110 7585043 109 + 24565398 ----UGGCAGACUUAUUCAGUCGAAUGCAUAAUAACCACGAUUAAAGCAAACAAGGCUCAUUGUCGGUUAAAUGACAAGUUUUGACAACUUGUGAAACGACAAGUGGUCAGUG------ ----..(((((((.....))))...)))....((((((((((...(((.......))).))))).)))))...........(((((.((((((......)))))).)))))..------ ( -25.50, z-score = -1.09, R) >consensus ___________CUAAGUCAGCCGCACGCAUAAUAACCACGAUUAAAGCAAACAAGGCUCAUUGUCGGUUAAAUGACAAGUUUUGACAACUUGUGAAACGACAAGUGGUCAGCGAUCG__ ...................((.(..(((....((((((((((...(((.......))).))))).)))))..(.(((((((.....))))))).)........)))..).))....... (-19.88 = -19.30 + -0.58)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:21:19 2011