| Sequence ID | dm3.chr3L |
|---|---|
| Location | 12,503,251 – 12,503,347 |
| Length | 96 |
| Max. P | 0.511301 |

| Location | 12,503,251 – 12,503,347 |
|---|---|
| Length | 96 |
| Sequences | 11 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 67.57 |
| Shannon entropy | 0.68354 |
| G+C content | 0.40648 |
| Mean single sequence MFE | -20.37 |
| Consensus MFE | -8.68 |
| Energy contribution | -8.15 |
| Covariance contribution | -0.53 |
| Combinations/Pair | 1.64 |
| Mean z-score | -0.78 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.511301 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
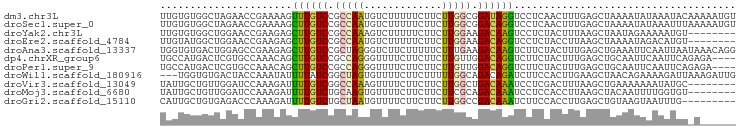
>dm3.chr3L 12503251 96 + 24543557 UUGUGUGGCUAGAACCGAAAAGUUUGUCCGCCAAUGUCUUUUUCUUCUUGGCGGAUAGGUCCUCAACUUUGAGCUAAAAUAUAAAUACAAAAAUGU ((((((((((.((((......))))(((((((((.............))))))))).))))((((....))))...........))))))...... ( -22.22, z-score = -1.84, R) >droSec1.super_0 4687219 96 + 21120651 UUGUGUGGCUAGAACCGAAAAGCUUGUCCGCCAAUGUCUUUUUCUUCUUGGCGGACAGGUCCUCAACUUUGAGCUAAAAUAUAAAUUUAAAAAUGU ((((((((((...........(((((((((((((.............)))))))))))))...........))))...))))))............ ( -26.57, z-score = -3.06, R) >droYak2.chr3L 12564059 88 + 24197627 UUGUGUGGCUGGAACCGAAGAGCUUGUCCGCCAAAGUCUUUUUCUUCUUGGAAGACAAGUCCUCUACUUUAAGCUAAUAGAAAAAUGU-------- .....(((((.(((....((..((((((..((((((.......))..))))..))))))..))....))).)))))............-------- ( -19.20, z-score = -0.36, R) >droEre2.scaffold_4784 12504540 88 + 25762168 UUGUAUGGCUGGAACCGAAGAGCUUGUCCGCCAAUGUCUUUUUCUUCUUGGAAGACAGGUCCUCUACCUUAAGCUAAAAUAGACAUGU-------- .....(((((........((..((((((..((((.............))))..))))))..))........)))))............-------- ( -18.51, z-score = 0.17, R) >droAna3.scaffold_13337 12552290 96 + 23293914 UGGUGUGACUGGAGCCGAAGAGCUUGUCCGCUAGGGUCUUCUUUUUCUUUGAAGACAAGUCUUCUACUUUGAGCUGAAAUUCAAUUAAUAAACAGG ........(((.....(((((.(((..((....))((((((.........))))))))))))))....(((((......)))))........))). ( -19.40, z-score = 1.11, R) >dp4.chrXR_group6 3285848 92 + 13314419 UGCCAUGACUCGUGCCAAACAGCUUGUCCGCCAGGGUUUUCUUCUUCUUGUUGGACAGGUCUUCUACUUUGAGCUGCAAUUCAAUUCAGAGA---- ((.((((...)))).))....(((((((((.(((((.........))))).)))))))))......(((((((.((.....)).))))))).---- ( -23.60, z-score = -0.32, R) >droPer1.super_9 1582407 92 + 3637205 UGCCAUGACUCGUGCCAAACAGCUUGUCCGCCAGGGUUUUCUUCUUCUUGUUGGACAGGUCUUCUACUUUGAGCUGCAAUUCAAUUCAGAGA---- ((.((((...)))).))....(((((((((.(((((.........))))).)))))))))......(((((((.((.....)).))))))).---- ( -23.60, z-score = -0.32, R) >droWil1.scaffold_180916 1100420 93 - 2700594 ---UGGUGUGACUACCAAAUAUUUUAUCGGCUAGUGUUUUCUUCUUUUUGGCAGACAGAUCUUCCACUUGAAGCUAACAGAAAAGAUUAAAGAUUG ---(((((....)))))........(((...((((.((((((.((((.(((.(((....))).)))...)))).....)))))).))))..))).. ( -15.40, z-score = 0.56, R) >droVir3.scaffold_13049 5901967 88 - 25233164 UAUUGCUGUUGGAUCCAAAGAUUUUGUCGGCCAAAGUUUUCUUCUUCUUGGCUGACAAAUCCUCGACUUUAAGCUGAAAAAAAUAUGC-------- ....(((...((.((...((..((((((((((((((.......))..))))))))))))..)).))))...)))..............-------- ( -21.60, z-score = -2.40, R) >droMoj3.scaffold_6680 17699398 88 - 24764193 UAUUGCUGUUGGAUCCAAAGAUUUUGUCUGCAAGUGUUUUCUUCUUCUUCGCAGACAAAUCCUCCACCUUAAGCUACAAUUUUGGUGU-------- ........(((....)))((..((((((((((((............))).)))))))))..)).((((..((.......))..)))).-------- ( -18.60, z-score = -1.44, R) >droGri2.scaffold_15110 7548721 87 - 24565398 CAUUGCUGUGAGACCCAAAGAUUUUGUCUGCUAAUGUUUUCUUCUUCUUGGCCGACAAAUCUUCCACCUUGAGCUGUAAGUAAUUUG--------- ....(((.((((.....(((((((.(((.(((((.............))))).))))))))))....))))))).............--------- ( -15.42, z-score = -0.64, R) >consensus UUGUGUGGCUGGAACCAAAGAGCUUGUCCGCCAAUGUUUUCUUCUUCUUGGCAGACAGGUCCUCUACUUUGAGCUAAAAUAAAAAUGU_A______ ......................((((((.(((((.............))))).))))))..................................... ( -8.68 = -8.15 + -0.53)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:21:15 2011