
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 12,489,508 – 12,489,642 |
| Length | 134 |
| Max. P | 0.538962 |

| Location | 12,489,508 – 12,489,642 |
|---|---|
| Length | 134 |
| Sequences | 5 |
| Columns | 151 |
| Reading direction | forward |
| Mean pairwise identity | 81.40 |
| Shannon entropy | 0.31936 |
| G+C content | 0.40416 |
| Mean single sequence MFE | -35.13 |
| Consensus MFE | -23.44 |
| Energy contribution | -25.02 |
| Covariance contribution | 1.58 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.538962 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 12489508 134 + 24543557 AAGCACAAUUAAAAGGCUCUAAACAAAAAACUAUACGAUUUUAGUUUGGCUACACCCACGCACCUAGUUUAGAGCAAUCAACAAAGGUGUGCAAGUGCUACACUGGGAGAAAUUC---AAGGAGCUAGCGAAAUAUA-------------- ..(((((........(((((((((...((((((........))))))((......)).........)))))))))............)))))...((((((.((.(((....)))---.))..).))))).......-------------- ( -31.85, z-score = -1.78, R) >droSim1.chr3L 11872645 134 + 22553184 AAGCACAAUUAAAAGGCUCUUAACUAAAAACCAUACGAUUUUCGUUUGGCUACACCCACGCACCUAGUUUAGAGCAAUCAACUAAGGUGUGCAAGUGCUACACUGGGAGAAAUUC---AAGGAGCUAGCAAAGUAUG-------------- ..............(((((((.........(((.(((.....))).))).....(((.(((((((((((..((....)))))).)))))))..((((...)))))))........---.)))))))...........-------------- ( -34.80, z-score = -1.92, R) >droSec1.super_0 4673366 147 + 21120651 AAGCACAAUUAAAAGGCUCUAAACUAAAAACUAUACGAUUUUCGUUUGGCUACACCCACGCACCUAGUUUAGAGCAAUCAACUAAGGUGUGCAAGUGCUACACUGGGAGAAAUUC---ACGGGGCUUA-GAAAUGGAGCUAGCAAAAUAUU ..(((((.(((....(((((((((((..........(.....)((.(((......))).))...))))))))))).......)))..)))))...((((...((((((....)))---.)))(((((.-......)))))))))....... ( -38.00, z-score = -2.10, R) >droYak2.chr3L 12550002 143 + 24197627 AAGCACAAUUAAAAGGCUCUAAA--AAA--CUAUACGAUUUUCGUUUGGCUACACCCACGCACCUAGUUUGGCGCAAUCUUCUAGGGUGCACAAGUGCUACACUGGGAAAAAUUC---AGGGAGCUGG-AAAAUGGAGUUAGCUAAAUAUG .(((.(........)))).....--...--.............(((((((((.(((((....((((((.((((((.((((....))))......)))))).)))))).....(((---((....))))-)...))).)))))))))))... ( -35.90, z-score = -1.05, R) >droEre2.scaffold_4784 12486183 147 + 25762168 AAGCACAAUUAAAAGGCUCUAAA--AAAAACUAUACAAUUUUCGUUUGGCUCCACCCUCGCACCUAGUUUAGUGCCAGCUUCUAAGGUGCUCAAGUGCUACACCGGGAAAAAUUCUUGAAAGAGCUUG-AAA-UGGAGCUUGCAAAAUAUG .(((.(........)))).....--............(((((.((..(((((((.....((((((((...(((....))).)).))))))((((((.((....(((((.....)))))..)).)))))-)..-))))))).)))))))... ( -35.10, z-score = -0.93, R) >consensus AAGCACAAUUAAAAGGCUCUAAAC_AAAAACUAUACGAUUUUCGUUUGGCUACACCCACGCACCUAGUUUAGAGCAAUCAACUAAGGUGUGCAAGUGCUACACUGGGAGAAAUUC___AAGGAGCUAG_AAAAUGGAG_U_GC_AAAUAU_ ..............((((((..........(((.(((.....))).))).....(((..((((((((((..((....)))))).))))))...((((...))))))).............))))))......................... (-23.44 = -25.02 + 1.58)

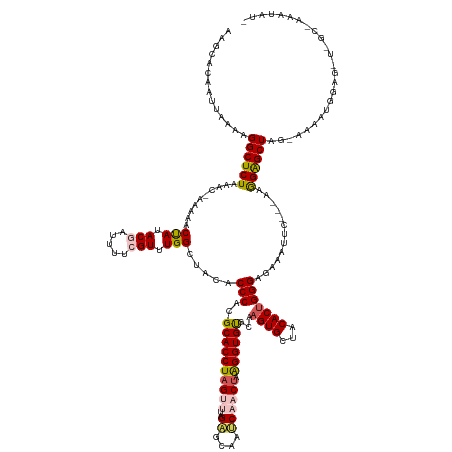
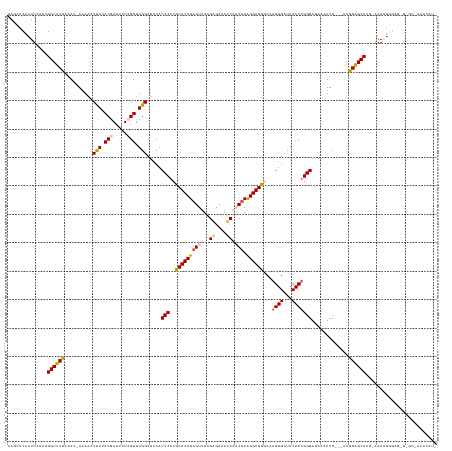
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:21:14 2011