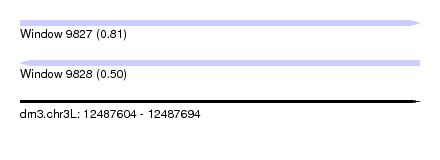
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 12,487,604 – 12,487,694 |
| Length | 90 |
| Max. P | 0.809957 |

| Location | 12,487,604 – 12,487,694 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 59.30 |
| Shannon entropy | 0.71905 |
| G+C content | 0.47480 |
| Mean single sequence MFE | -19.85 |
| Consensus MFE | -6.13 |
| Energy contribution | -6.09 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.31 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.809957 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
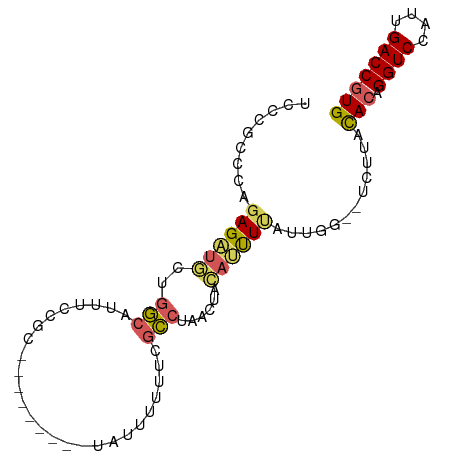
>dm3.chr3L 12487604 90 + 24543557 UCCCGUCCAGAGAUGCUGGCAUUUCCGCGUAUUUUUUUUUUUUUUCGCCUAACCACACUUUAUUGG--UCUUACACAGGUCCAUUGACCGUG ....(.(((((((((....)))))).(((................)))...............)))--.)...(((.((((....))))))) ( -16.69, z-score = -1.25, R) >droYak2.chr3L 12547888 83 + 24197627 UCCCGCCCAGAGAUGCUGGCAUUUCCGC---------UAUUUUUUCGCCUUACUACAUUUCGUCGGGUCUUUACACAGGUCCAUUGACCGUG .((((....((((((..(((........---------.........)))......))))))..))))......(((.((((....))))))) ( -19.33, z-score = -1.37, R) >droEre2.scaffold_4784 12484220 81 + 25762168 UCUCGCCCAGAGAUGCUGGCAUUUCCGC---------UAUUUUUUCGCCUCACUACAUUUCAUUGG--UCUUACACAGGUCCAUUGACCGUG ....(((..((((((..(((........---------.........)))......))))))...))--)....(((.((((....))))))) ( -14.93, z-score = -0.68, R) >droWil1.scaffold_180916 1083304 85 - 2700594 UUUUCGUCAGAGUUUGCUCCAUUUUCAUAG------CUAAACGAUUAGCUAUUUAGAGUUUGCCAG-CUGUUUCACCGGUCCAUUGACCGUG .....(..(.((((.((....(((..((((------((((....))))))))..)))....)).))-)).)..)..(((((....))))).. ( -20.90, z-score = -2.78, R) >droMoj3.scaffold_6680 6083479 81 + 24764193 ------CCAUAGACAGAGCCACAUCCACAGGUGA---UCGGCUGCUGGCUGUUGGCUUCUUUUUGU--CUAUCUACCGGUCCAUUGACCAUG ------..(((((((((((((((.((((((.(..---..).))).))).)).)))))).....)))--)))).....((((....))))... ( -27.40, z-score = -2.44, R) >consensus UCCCGCCCAGAGAUGCUGGCAUUUCCGC_________UAUUUUUUCGCCUAACUACAUUUUAUUGG__UCUUACACAGGUCCAUUGACCGUG .........((((((..(((..........................)))......))))))............(((.((((....))))))) ( -6.13 = -6.09 + -0.04)

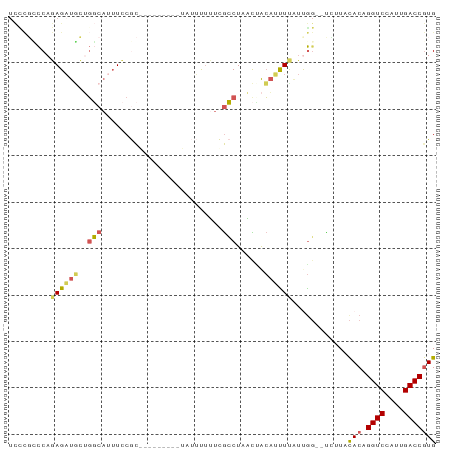
| Location | 12,487,604 – 12,487,694 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 59.30 |
| Shannon entropy | 0.71905 |
| G+C content | 0.47480 |
| Mean single sequence MFE | -20.64 |
| Consensus MFE | -8.04 |
| Energy contribution | -7.40 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.36 |
| Mean z-score | -0.86 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
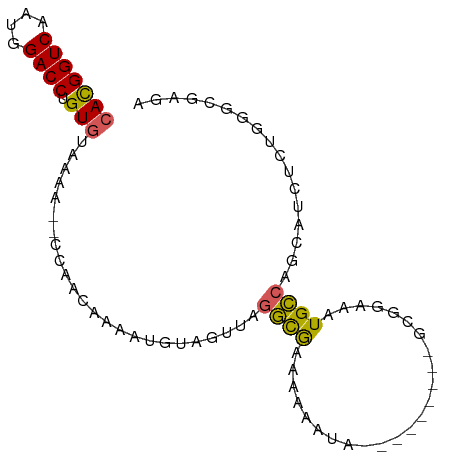
>dm3.chr3L 12487604 90 - 24543557 CACGGUCAAUGGACCUGUGUAAGA--CCAAUAAAGUGUGGUUAGGCGAAAAAAAAAAAAAAUACGCGGAAAUGCCAGCAUCUCUGGACGGGA .............(((((((..((--(((........)))))..))...................((((.(((....))).)))).))))). ( -18.40, z-score = -0.18, R) >droYak2.chr3L 12547888 83 - 24197627 CACGGUCAAUGGACCUGUGUAAAGACCCGACGAAAUGUAGUAAGGCGAAAAAAUA---------GCGGAAAUGCCAGCAUCUCUGGGCGGGA (((((((....)))).)))......((((.(((.((((......((.........---------))((.....)).)))).)).)..)))). ( -20.20, z-score = -0.15, R) >droEre2.scaffold_4784 12484220 81 - 25762168 CACGGUCAAUGGACCUGUGUAAGA--CCAAUGAAAUGUAGUGAGGCGAAAAAAUA---------GCGGAAAUGCCAGCAUCUCUGGGCGAGA (((((((....)))).)))...(.--((...((.((((......((.........---------))((.....)).)))).))..))).... ( -15.70, z-score = 0.65, R) >droWil1.scaffold_180916 1083304 85 + 2700594 CACGGUCAAUGGACCGGUGAAACAG-CUGGCAAACUCUAAAUAGCUAAUCGUUUAG------CUAUGAAAAUGGAGCAAACUCUGACGAAAA ....((((..((.(((((......)-)))))...(((((.((((((((....))))------)))).....)))))....)..))))..... ( -22.50, z-score = -2.85, R) >droMoj3.scaffold_6680 6083479 81 - 24764193 CAUGGUCAAUGGACCGGUAGAUAG--ACAAAAAGAAGCCAACAGCCAGCAGCCGA---UCACCUGUGGAUGUGGCUCUGUCUAUGG------ ..(((((....)))))((((((((--(.........((((.((.(((.(((....---....)))))).)))))))))))))))..------ ( -26.40, z-score = -1.78, R) >consensus CACGGUCAAUGGACCUGUGUAAAA__CCAACAAAAUGUAGUUAGGCGAAAAAAUA_________GCGGAAAUGCCAGCAUCUCUGGGCGAGA (((((((....)))).)))........................((((........................))))................. ( -8.04 = -7.40 + -0.64)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:21:12 2011