| Sequence ID | dm3.chr3L |
|---|---|
| Location | 12,407,626 – 12,407,698 |
| Length | 72 |
| Max. P | 0.822608 |

| Location | 12,407,626 – 12,407,698 |
|---|---|
| Length | 72 |
| Sequences | 10 |
| Columns | 79 |
| Reading direction | forward |
| Mean pairwise identity | 70.38 |
| Shannon entropy | 0.61425 |
| G+C content | 0.35792 |
| Mean single sequence MFE | -11.37 |
| Consensus MFE | -6.38 |
| Energy contribution | -6.38 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.85 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.822608 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
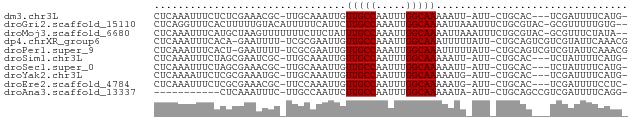
>dm3.chr3L 12407626 72 + 24543557 CUCAAAUUUCUCUCGAAACGC-UUGCAAAUUGUUGCCAAUUUGGCAAAAAUU-AUU-CUGCAC---UCGAUUUUCAUG- ..............((((((.-.((((((((.(((((.....))))).))))-...-.)))).---.))..))))...- ( -13.80, z-score = -2.38, R) >droGri2.scaffold_15110 19448607 76 - 24565398 CUCAGGUUUCACUUUUUGUACAUUUUUCAUUCUUGCCAAAUUGGCAAAAUUAAAUUUCUGCGUAC-GCGUUUUUGUG-- .................(((.(..(((.(((.(((((.....)))))))).)))..).)))...(-(((....))))-- ( -9.30, z-score = -0.04, R) >droMoj3.scaffold_6680 6012412 76 + 24764193 CUCAAAUUUCAUGCUAAGUUUUUUUCUUCUAUUUGCCAAAUUGGCAAAAUUAAAUUUCUGCGUAC-GCGUUUCUAUA-- ..........((((.((((((..((.......(((((.....)))))))..))))))..))))..-...........-- ( -9.71, z-score = -1.01, R) >dp4.chrXR_group6 3189170 76 + 13314419 CUCAAAUUUCACA-GAAUUUU-UCGCGAAUUGUUGCCAAAUUGGCAAAUUUUUAUU-CUGCAGUCGUCGUAUUCAAACG .............-((((...-.(((((.((((((((.....))))..........-..))))))).)).))))..... ( -12.20, z-score = -0.78, R) >droPer1.super_9 1483774 76 + 3637205 CUCAAAUUUCACU-GAAUUUU-UCGCGAAUUGUUGCCAAAUUGGCAAAUUUUUAUU-CUGCAGUCGUCGUAUUCAAACG ............(-((((...-.(((((.((((((((.....))))..........-..))))))).)).))))).... ( -12.80, z-score = -0.97, R) >droSim1.chr3L 11788712 72 + 22553184 CUCAAAUUUCUAGCGAAUCGC-UUGCAAAUUGUUGCCAAUUUGGCAAAAAUU-AUU-CUGCAC---UCUAUUUUCAUG- ...........((((...)))-)((((((((.(((((.....))))).))))-...-.)))).---............- ( -12.30, z-score = -1.29, R) >droSec1.super_0 4591612 72 + 21120651 CUCAAAUUUCUAGCGAAACGC-UUGCAAAUUGUUGCCAAUUUGGCAAAAAUU-AUU-CUGCAC---UCUAUUUUCAUG- ...........((((...)))-)((((((((.(((((.....))))).))))-...-.)))).---............- ( -12.30, z-score = -1.28, R) >droYak2.chr3L 12463558 72 + 24197627 CUCAAAAUUCUCGCGAAAUGC-UUGCAAAUUGUUGCCAAUUUGGCAAAAAUG-AUU-CUGCAC---UCGAUUUUCAUG- ..............((((((.-.((((((((.(((((.....)))))....)-)))-.)))).---.))..))))...- ( -12.50, z-score = -0.38, R) >droEre2.scaffold_4784 12402495 72 + 25762168 CUCAAAUUUCUCGCGAAACGC-UUCCAAAUUGUUGCCAAUUUGGCAAAAAUG-AUU-CUGCAC---UCGAUUUUCCUC- .............(((...((-......(((.(((((.....))))).))).-...-..))..---))).........- ( -11.10, z-score = -0.90, R) >droAna3.scaffold_13337 22045506 64 - 23293914 -----------CUCAAAUUUC-UUGCCAAUUCUUGCCAAUUUGGCAAAAAUA-AUU-CUGCAGCCGUCGAUUUUCAGG- -----------.....((((.-(((((((...........))))))))))).-...-.....................- ( -7.70, z-score = 0.52, R) >consensus CUCAAAUUUCUCGCGAAAUGC_UUGCAAAUUGUUGCCAAUUUGGCAAAAAUU_AUU_CUGCAC___UCGAUUUUCAUG_ ................................(((((.....)))))................................ ( -6.38 = -6.38 + -0.00)


| Location | 12,407,626 – 12,407,698 |
|---|---|
| Length | 72 |
| Sequences | 10 |
| Columns | 79 |
| Reading direction | reverse |
| Mean pairwise identity | 70.38 |
| Shannon entropy | 0.61425 |
| G+C content | 0.35792 |
| Mean single sequence MFE | -15.01 |
| Consensus MFE | -6.19 |
| Energy contribution | -6.19 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.607002 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr3L 12407626 72 - 24543557 -CAUGAAAAUCGA---GUGCAG-AAU-AAUUUUUGCCAAAUUGGCAACAAUUUGCAA-GCGUUUCGAGAGAAAUUUGAG -........((((---((((((-((.-...))))))(((((((....)))))))...-))(((((....))))))))). ( -18.60, z-score = -2.39, R) >droGri2.scaffold_15110 19448607 76 + 24565398 --CACAAAAACGC-GUACGCAGAAAUUUAAUUUUGCCAAUUUGGCAAGAAUGAAAAAUGUACAAAAAGUGAAACCUGAG --(((......((-....)).....((((.(((((((.....))))))).)))).............)))......... ( -14.40, z-score = -2.23, R) >droMoj3.scaffold_6680 6012412 76 - 24764193 --UAUAGAAACGC-GUACGCAGAAAUUUAAUUUUGCCAAUUUGGCAAAUAGAAGAAAAAAACUUAGCAUGAAAUUUGAG --.........((-....)).....(((...((((((.....))))))...))).......(((((........))))) ( -9.20, z-score = -0.15, R) >dp4.chrXR_group6 3189170 76 - 13314419 CGUUUGAAUACGACGACUGCAG-AAUAAAAAUUUGCCAAUUUGGCAACAAUUCGCGA-AAAAUUC-UGUGAAAUUUGAG ((((.......))))...((((-(((.....(((((....(((....)))...))))-)..))))-))).......... ( -14.20, z-score = -1.00, R) >droPer1.super_9 1483774 76 - 3637205 CGUUUGAAUACGACGACUGCAG-AAUAAAAAUUUGCCAAUUUGGCAACAAUUCGCGA-AAAAUUC-AGUGAAAUUUGAG ((((.......))))....(((-(........(((((.....)))))...(((((..-.......-.))))).)))).. ( -12.10, z-score = -0.09, R) >droSim1.chr3L 11788712 72 - 22553184 -CAUGAAAAUAGA---GUGCAG-AAU-AAUUUUUGCCAAAUUGGCAACAAUUUGCAA-GCGAUUCGCUAGAAAUUUGAG -............---..((((-((.-...))))))(((((((....)))))))..(-(((...))))........... ( -15.30, z-score = -1.35, R) >droSec1.super_0 4591612 72 - 21120651 -CAUGAAAAUAGA---GUGCAG-AAU-AAUUUUUGCCAAAUUGGCAACAAUUUGCAA-GCGUUUCGCUAGAAAUUUGAG -...((((...(.---.((((.-...-((((.(((((.....))))).)))))))).-.).)))).............. ( -15.30, z-score = -1.13, R) >droYak2.chr3L 12463558 72 - 24197627 -CAUGAAAAUCGA---GUGCAG-AAU-CAUUUUUGCCAAAUUGGCAACAAUUUGCAA-GCAUUUCGCGAGAAUUUUGAG -........((((---((((((-((.-...))))))(((((((....)))))))...-))..(((....)))..)))). ( -17.20, z-score = -1.11, R) >droEre2.scaffold_4784 12402495 72 - 25762168 -GAGGAAAAUCGA---GUGCAG-AAU-CAUUUUUGCCAAAUUGGCAACAAUUUGGAA-GCGUUUCGCGAGAAAUUUGAG -........((((---((((((-((.-...))))))(((((((....)))))))...-))(((((....))))))))). ( -20.00, z-score = -2.20, R) >droAna3.scaffold_13337 22045506 64 + 23293914 -CCUGAAAAUCGACGGCUGCAG-AAU-UAUUUUUGCCAAAUUGGCAAGAAUUGGCAA-GAAAUUUGAG----------- -........((((...((((..-...-.(((((((((.....)))))))))..)).)-)....)))).----------- ( -13.80, z-score = -1.03, R) >consensus _CAUGAAAAUCGA___GUGCAG_AAU_AAUUUUUGCCAAAUUGGCAACAAUUUGCAA_GCAAUUCGCGAGAAAUUUGAG ................................(((((.....)))))................................ ( -6.19 = -6.19 + -0.00)

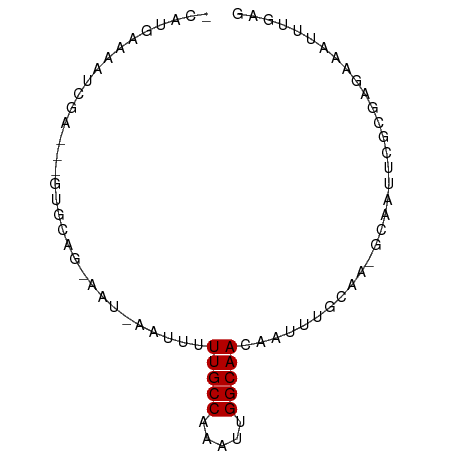
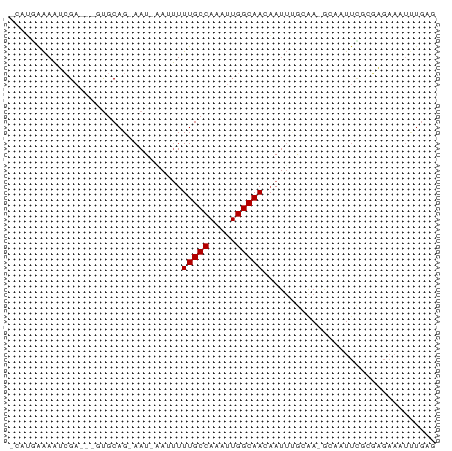
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:21:03 2011