| Sequence ID | dm3.chr3L |
|---|---|
| Location | 12,404,785 – 12,404,889 |
| Length | 104 |
| Max. P | 0.646195 |

| Location | 12,404,785 – 12,404,889 |
|---|---|
| Length | 104 |
| Sequences | 12 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 67.63 |
| Shannon entropy | 0.70100 |
| G+C content | 0.39193 |
| Mean single sequence MFE | -14.26 |
| Consensus MFE | -7.60 |
| Energy contribution | -8.19 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.08 |
| Mean z-score | -0.37 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.646195 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
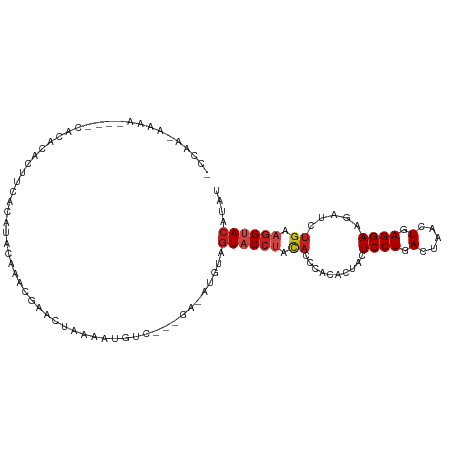
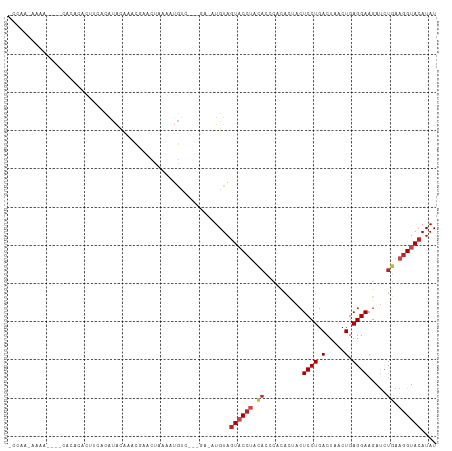
>dm3.chr3L 12404785 104 - 24543557 ACCGAAAAAA----CACACACUUCACACACUAACAAACCAAAAUGUC---GA-AUGUAGUACCUACACCCACACUACUCCUGACUAACUGAGGAAGAUCUGAAGGUACAUAU ..........----...((.(((((..................(((.---(.-.(((((...)))))..))))....((((.(.....).)))).....)))))))...... ( -11.70, z-score = 0.28, R) >droGri2.scaffold_15110 19445402 87 + 24565398 -------------------CCUAUUUAUAUCCGCGUACUAUAAUGCUUAUAUUGUGUAGUACCUGCA------UAACUCCUGACUAACUGAGGAUGAUUUGAAGGUACAUAU -------------------..(((..((((..((((......))))..))))...)))((((((.((------....((((.(.....).)))).....)).)))))).... ( -17.00, z-score = -1.11, R) >droMoj3.scaffold_6680 6009578 102 - 24764193 ---------CUACCUAUAUACAAAACAUAUAUGUGUACUAUAUUGCUUU-AAAAUGUAGUUCCUGCAUACACAUCACUCCUGACUAACUGAGGAAGACUUGAAGGUACAUAU ---------.(((((..............((((((.((((((((.....-..))))))))...))))))((..((..((((.(.....).)))).))..)).)))))..... ( -17.70, z-score = -0.69, R) >droVir3.scaffold_13049 7762580 92 - 25233164 -------UUCUUCCUAUAUACUUAACAUACCUGUGUACUAUAUUGCUUA-AAAAUGUAGUUC------------CAGUCCUGACUAGCUGAGGAAGACUUGAAGGUACAUAU -------.(((((((.....((...((...(((.(.((((((((.....-..)))))))).)------------)))...))...))...)))))))............... ( -18.10, z-score = -0.77, R) >droWil1.scaffold_180727 1441451 95 + 2741493 --------------AUCAACUUACGCGUGCUACAUACCUAAAAAAUG---AACAUGUAGUACCCCUACACACAUCACUCCUGACUAACUGAGGAAGAUCUGAAGGUAGAUAU --------------............(((((((((..(........)---...)))))))))..((((.((.(((..((((.(.....).)))).))).))...)))).... ( -18.10, z-score = -1.64, R) >dp4.chrXR_group6 3186617 105 - 13314419 -CAAAAAAACACUCCACAUAA-UCUCGUACAUACGCUCUAAACUGCC----U-CUGUAGUACCUCCACACUUACAACUCCUGACUAACUGAGGAAGAUCUGAAGGUACAUAU -....................-.....((((...((........)).----.-.))))((((((.((..(((....(((..........))).)))...)).)))))).... ( -14.20, z-score = -0.62, R) >droPer1.super_9 1481205 105 - 3637205 -CAAACAAACACUCCACAUAA-UCUCGUACAUACGCUCUAAACUGCC----U-CUGUAGUACCUCCACACUUACAACUCCUGACUAACUGAGGAAGAUCUGAAGGUACAUAU -....................-.....((((...((........)).----.-.))))((((((.((..(((....(((..........))).)))...)).)))))).... ( -14.20, z-score = -0.44, R) >droAna3.scaffold_13337 22043049 99 + 23293914 ACAUACAA--------CACCACCCACAUACGCUCCAAAACCAAUGUC----A-AUGUAGUACCUACACCCACACUACUCCUGACUAACUGAGGAAGAUCUGAAGGUACAUAU ........--------....(((..((.....(((.........(((----(-..(((((............)))))...)))).......))).....))..)))...... ( -11.09, z-score = 0.59, R) >droEre2.scaffold_4784 12399687 101 - 25762168 ACCAACAAAA-------ACAUCUCACACACGAACUAACCAAAAUGUC---GA-AUGUAGUACCUACACCCACACUACUCCUGACUAACUGAGGAAGAUCUGAAGGUACAUAU ..........-------.......(((..(((.(..........)))---).-.))).((((((.((......((..((((.(.....).))))))...)).)))))).... ( -12.10, z-score = 0.17, R) >droYak2.chr3L 12460708 101 - 24197627 ACCAACAAAA-------ACACCUCACACACAAACUAACCAAAAUGUC---GA-UUGUAGUACCUACACCCACACUACUCCUGACUAACUGAGGAAGAUCUGAAGGUACAUAU (((..((...-------...(((((...................(((---(.-..(((((............)))))...))))....)))))......))..)))...... ( -12.80, z-score = -0.23, R) >droSec1.super_0 4588734 108 - 21120651 ACCAAAAAAACAAACACACACUUCACACACUAACUAACCAAAAUGUC---GA-AUGUAGUACCUACACCCACACUACUCCUGACUAACUGAGGAAGAUCUGAAGGUACAUAU .................((.(((((....((..((.........(((---(.-..(((((............)))))...))))......))..))...)))))))...... ( -12.46, z-score = -0.16, R) >droSim1.chr3L 11785824 108 - 22553184 ACCAAAAAAACAAACACACACUUCACACACCAACUAACCAAAAUGUC---GA-AUGUAGUACCUACACCCACACUACUCCUGACUAACUGAGGAAGAUCUGAAGGUACAUAU .................((.(((((..................(((.---(.-.(((((...)))))..))))....((((.(.....).)))).....)))))))...... ( -11.70, z-score = 0.15, R) >consensus _CCAA_AAAA____CACACACUUCACAUACAAACGAACUAAAAUGUC___GA_AUGUAGUACCUACACCCACACUACUCCUGACUAACUGAGGAAGAUCUGAAGGUACAUAU ..........................................................((((((.((..........((((.(.....).)))).....)).)))))).... ( -7.60 = -8.19 + 0.60)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:21:01 2011