
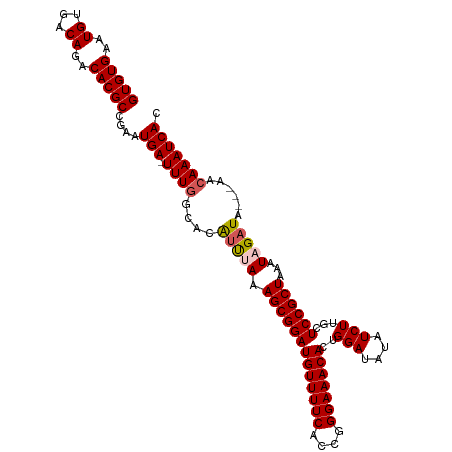
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 12,399,323 – 12,399,488 |
| Length | 165 |
| Max. P | 0.798740 |

| Location | 12,399,323 – 12,399,432 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 94.80 |
| Shannon entropy | 0.08781 |
| G+C content | 0.41570 |
| Mean single sequence MFE | -26.13 |
| Consensus MFE | -23.28 |
| Energy contribution | -23.24 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.89 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.798740 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
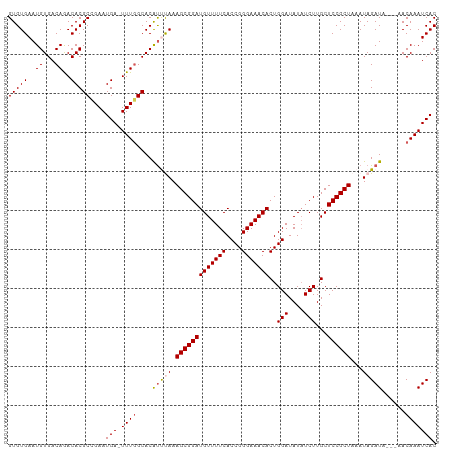
>dm3.chr3L 12399323 109 - 24543557 GUGUGAAUGUGACAGACACGCCGAAUGA-UUUGGCACAUUCAAAGCGGAUGUUUUCACCGGGAAACACUGGAUAUAUCUUGCUCCGCUAAAUAGAUAAACAACAAAUCAC (((((((((((.((((((.......)).-)))).)))))))).((((((.((.....((((......)))).........))))))))...................))) ( -25.94, z-score = -1.51, R) >droEre2.scaffold_4784 12394137 106 - 25762168 GUGUGAAUGUGACAAACACGCAGAAUGA-UUUGGCACAUCUAAAGCGGAUGUUUUCACCGGGAAACACUGGAUAUAUCUUGCUCCGCUAAAUAGAUA---AACAAAUCAC (((((...........)))))....(((-((((....(((((.((((((.((.....((((......)))).........))))))))...))))).---..))))))). ( -28.04, z-score = -2.78, R) >droYak2.chr3L 12455001 107 - 24197627 GUGUGAAUGUGACAAACACGCAGAAUGAUUUUUGCACGCCUAAAGCGGAUGUUUUCACCGGGAAACACUGGAUAUUUCUUGCUCCGCUAAAUAGAUA---AACAAAUCAC ((((....(((.....)))((((((....))))))))))....(((((((((((((....)))))))..(((....)))...)))))).....(((.---.....))).. ( -25.10, z-score = -1.54, R) >droSec1.super_0 4583260 106 - 21120651 GUGUGAAUGUGACAGACACGCCGAAUGA-UUUGGCACAUUUAAAGCGGAUGUUUUCACCGGGAAACACUGGAUAUAUCUUGCUCCGCUAAAUAGAUA---AACAAAUCAC (((((..((...))..)))))....(((-((((....(((((.((((((.((.....((((......)))).........))))))))...))))).---..))))))). ( -25.54, z-score = -1.64, R) >droSim1.chr3L 11780189 106 - 22553184 GUGUGAAUGUGACAGACACGCCGAAUGA-UUUGGCACAUUCAAAGCGGAUGUUUUCACCGGGAAACACUGGAUAUAUCUUGCUCCGCUAAAUAGAUA---AACAAAUCAC ...((((((((.((((((.......)).-)))).)))))))).((((((.((.....((((......)))).........)))))))).....(((.---.....))).. ( -26.04, z-score = -1.56, R) >consensus GUGUGAAUGUGACAGACACGCCGAAUGA_UUUGGCACAUUUAAAGCGGAUGUUUUCACCGGGAAACACUGGAUAUAUCUUGCUCCGCUAAAUAGAUA___AACAAAUCAC ..((((..(((.....)))((((((....))))))........(((((((((((((....)))))))..(((....)))...))))))..................)))) (-23.28 = -23.24 + -0.04)

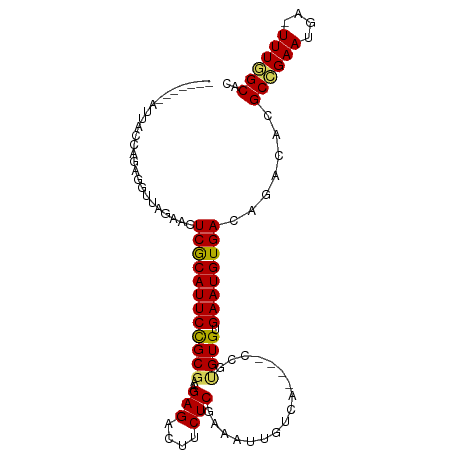
| Location | 12,399,396 – 12,399,488 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 81.33 |
| Shannon entropy | 0.30862 |
| G+C content | 0.46279 |
| Mean single sequence MFE | -26.92 |
| Consensus MFE | -18.30 |
| Energy contribution | -18.34 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.788336 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
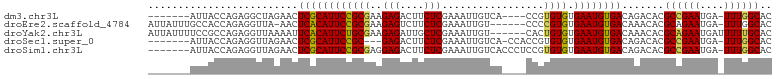
>dm3.chr3L 12399396 92 - 24543557 -------AUUACCAGAGGCUAGAACUCGCAUUCCGCGAAGAGACUUCUCGAAAUUGUCA----CCGUGUGUGAAUGUGACAGACACGCCGAAUGA-UUUGGCAC -------..........((((((.....(((((.(((..(((....)))....((((((----(.((......)))))))))...))).))))).-)))))).. ( -25.50, z-score = -1.33, R) >droEre2.scaffold_4784 12394207 96 - 25762168 AUUAUUUGCCACCAGAGGUUA-AACUCACAUUCCGCGAAGAGUCUUCUCGAAAUUGU------CCCCGUGUGAAUGUGACAAACACGCAGAAUGA-UUUGGCAC ......(((((...(((....-..))).(((((.(((..(((....)))....((((------(..(((....))).)))))...))).))))).-..))))). ( -27.50, z-score = -2.60, R) >droYak2.chr3L 12455071 98 - 24197627 AUUAUUUUCCGCCAGAGGUUAAAAUUCACAUUCUGCGAAGAGAUUGCUCGAAAUUGU------CACUGUGUGAAUGUGACAAACACGCAGAAUGAUUUUUGCAC ..........((..(((.......))).(((((((((..(((....)))....((((------(((.........)))))))...)))))))))......)).. ( -26.80, z-score = -2.21, R) >droSec1.super_0 4583330 92 - 21120651 -------AUUACCAGAGGUUAGAACUCGCAUUCCGC---GAGACUUCUCGAAAUUGUCA-CCACCGUGUGUGAAUGUGACAGACACGCCGAAUGA-UUUGGCAC -------....(.((((((.....(((((.....))---))))))))).)...((((((-((((.....)))...)))))))....(((((....-.))))).. ( -26.20, z-score = -1.61, R) >droSim1.chr3L 11780259 96 - 22553184 -------AUUACCAGAGGUUAGAACUCGCAUUCCGCGAGGAGACUUCUCGAAAUUGUCACCCUCCGUGUGUGAAUGUGACAGACACGCCGAAUGA-UUUGGCAC -------....(.(((((((....(((((.....)))))..))))))).)...(((((((..(((....).))..)))))))....(((((....-.))))).. ( -28.60, z-score = -1.64, R) >consensus _______AUUACCAGAGGUUAGAACUCGCAUUCCGCGAAGAGACUUCUCGAAAUUGUCA____CCGUGUGUGAAUGUGACAGACACGCCGAAUGA_UUUGGCAC .........................((((((((((((..(((....)))((.....))........)))).)))))))).......((((((....)))))).. (-18.30 = -18.34 + 0.04)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:21:00 2011