
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 12,383,658 – 12,383,767 |
| Length | 109 |
| Max. P | 0.662975 |

| Location | 12,383,658 – 12,383,767 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 66.91 |
| Shannon entropy | 0.57388 |
| G+C content | 0.39214 |
| Mean single sequence MFE | -27.28 |
| Consensus MFE | -11.64 |
| Energy contribution | -11.37 |
| Covariance contribution | -0.27 |
| Combinations/Pair | 1.57 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.662975 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 12383658 109 - 24543557 ---UUCGUUUUCGUAUCGCACUCUUGCGGGUAAAUAUAUGUGGUAUUUACCAAAGUAAACUACAUCGUGUG-----CGUGUUGCAGUGAGAAG---UGCAUUUAUUGCUGCAAAUUCGUA ---...((((.(....((((....))))(((((((((.....)))))))))...).)))).........((-----((..((((((..((((.---....))).)..))))))...)))) ( -28.10, z-score = -0.91, R) >droAna3.scaffold_13337 22023739 105 + 23293914 ----UCAGUUUCGGAUU------UCUCUGGCGCG-GUAUUUGGUCUUUACCCAACAGCGGUAUAAAGUGUAAAUUUUGUGUGCCAGUGAAAAG---UGAAUAAUUUUGAGGCGAUUGGG- ----...((((((((((------((.((((((((-(((.........))))..........(((((((....)))))))))))))).))))..---........)))))))).......- ( -26.60, z-score = -0.92, R) >droEre2.scaffold_4784 12378684 97 - 25762168 UUCCACGUUGCCGAAUC------UCACUGGCGCGGGCA---AAUAUUUACCAAAGUGAACUA------GUG-----CGUGUUGCAGUGAGAAG---UGCAUUAAUUACUGCAAAUUCGGA ..........(((((((------((((((..(((.(((---....(((((....)))))...------.))-----).)))..))))))))..---((((........))))..))))). ( -31.10, z-score = -2.13, R) >droYak2.chr3L 12438999 94 - 24197627 ---UACGUUGCCGUAUC------UCACUGGUGCGGGUA---AAUGUUUACCACAGUGAACUA------GUG-----CGUGUUGCAGUGAGAAG---UGCAUUAAUUACUGCAUAUUCGGA ---.......(((..((------((((((..(((.(((---...((((((....))))))..------.))-----).)))..)))))))).(---((((........)))))...))). ( -31.90, z-score = -2.20, R) >droSec1.super_0 4567566 108 - 21120651 ---UUCCUUUUCGAAUC----UAUUGCGGGUAAAUAUAUCUAGUAUUUACCAAAGUGAACUACAUAGUGUG-----CGUGUUGCAGUGAUAAUUUUUGCAUUAAUUGCUGCAAAUUCAAA ---.........((((.----....((((((((((((.....)))))))))..((....))........))-----)...((((((..(((((......))).))..))))))))))... ( -23.80, z-score = -1.51, R) >droSim1.chr3L 11764535 108 - 22553184 ---UUCCUUUUCGAAUC----UAUUGCGGGUAAAUAUAUGUAAUAAUUACCAAAGUGAACUACAUAUUGUG-----CGUGUUGCAGUGAGAAUUUUUGCAUUAAUUGCUGCAAAUUUGAA ---......(((((((.----....(((.(((...(((((((....((((....))))..)))))))..))-----).)))(((((..(...............)..))))).))))))) ( -22.16, z-score = -0.82, R) >consensus ___UUCGUUUUCGAAUC______UCGCGGGUAAAUAUAU_UAAUAUUUACCAAAGUGAACUACAUAGUGUG_____CGUGUUGCAGUGAGAAG___UGCAUUAAUUGCUGCAAAUUCGGA ..........((((((.......((((.((((...............))))...))))......................(((((((((...............))))))))))))))). (-11.64 = -11.37 + -0.27)

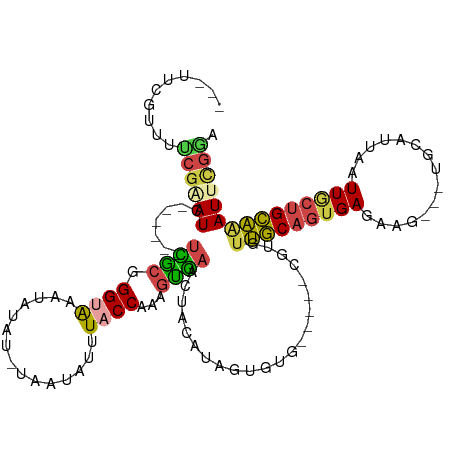
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:20:58 2011