
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 12,378,149 – 12,378,279 |
| Length | 130 |
| Max. P | 0.698872 |

| Location | 12,378,149 – 12,378,279 |
|---|---|
| Length | 130 |
| Sequences | 12 |
| Columns | 152 |
| Reading direction | forward |
| Mean pairwise identity | 69.32 |
| Shannon entropy | 0.61865 |
| G+C content | 0.45024 |
| Mean single sequence MFE | -36.81 |
| Consensus MFE | -12.64 |
| Energy contribution | -13.24 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.34 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.698872 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
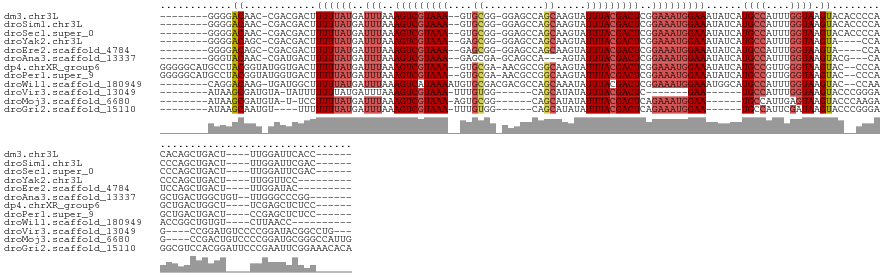
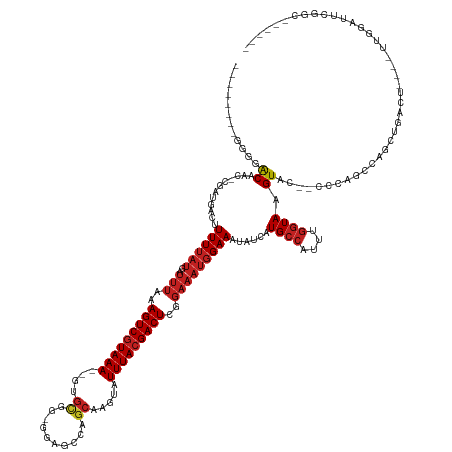
>dm3.chr3L 12378149 130 + 24543557 --------GGGGACAAC-CGACGACUUUUUAUGAUUUAAAGUCGUAAA--GUGCGG-GGAGCCAGCAAGUAUUUACGACUCGGAAAUGGAAAUAUCAUGCCAUUUGGUAAGUACACCCCACACAGCUGACU----UUGGAUUCACC------ --------(....)..(-((((((((((.........))))))))...--(((.((-((.(....)..((((((((.(...((..((((.....)))).))...).)))))))).)))).)))........----.))).......------ ( -33.80, z-score = -1.02, R) >droSim1.chr3L 11758689 130 + 22553184 --------GGGGACAAC-CGACGACUUUUUAUGAUUUAAAGUCGUAAA--GUGCGG-GGAGCCAGCAAGUAUUUACGACUCGGAAAUGGAAAUAUCAUGCCAUUUGGUAAGUACACCCCACCCAGCUGACU----UUGGAUUCGAC------ --------((((.....-....((..((((....(((..(((((((((--.(((((-....)).)))....)))))))))..)))...))))..)).((((....))))......))))..((((......----)))).......------ ( -35.00, z-score = -1.11, R) >droSec1.super_0 4562003 130 + 21120651 --------GGGGACAAC-CGACGACUUUUUAUGAUUUAAAGUCGUAAA--GUGCGG-GGAGCCAGCAAGUAUUUACGACUCGGAAAUGGAAAUAUCAUGCCAUUUGGUAAGUACACCCCACCCAGCUGACU----UUGGAUUCGAC------ --------((((.....-....((..((((....(((..(((((((((--.(((((-....)).)))....)))))))))..)))...))))..)).((((....))))......))))..((((......----)))).......------ ( -35.00, z-score = -1.11, R) >droYak2.chr3L 12433506 123 + 24197627 --------GGGGACAGC-CGACGACUUUUUAUGAUUUAAAGUCGUAAA--GAGCGG-GGAGCCAGCAAGUAUUUACGACUCGGAAAUGGAAAUAUCAUGCCAUUUGGUAAGUA----CCACCCAGCUGACU----UUGGUUCC--------- --------.(((((.((-(..((.(((((((((((.....))))))))--))))).-.).))((((..((....)).....((((((((..........))))))(((....)----))..)).))))...----...)))))--------- ( -34.10, z-score = -1.27, R) >droEre2.scaffold_4784 12373316 123 + 25762168 --------GGGGACAGC-CGACGACUUUUUAUGAUUUAAAGUCGUAAA--GAGCGG-GGAGCCAGCAAGUAUUUACGACUCGGAAAUGGAAAUAUCAUGCCAUUUGGUAAGUA----CCAUCCAGCUGACU----UUGGAUAC--------- --------..((....(-(..((.(((((((((((.....))))))))--))))).-))..)).....(((((((.((.((((...((((.....(.((((....)))).)..----...)))).)))).)----))))))))--------- ( -33.30, z-score = -1.67, R) >droAna3.scaffold_13337 22018889 125 - 23293914 --------GGGUACAAC-CGAUGACUUUUUAUGAUUUAAAGUCGUAAA--GAGCGA-GCAGCCA---AGUAUUUACGACUCGGAAAUGGAAAUAUCAUGCCAUUUGGUAAGUACG---CAGCUGACUGGCUGU--UUGGGCCCGG------- --------((((.....-......(((((((((((.....))))))))--)))(((-(((((((---.((((((((.(...((..((((.....)))).))...).)))))))).---(....)..)))))))--))).))))..------- ( -39.80, z-score = -2.60, R) >dp4.chrXR_group6 3158643 137 + 13314419 GGGGGCAUGCCUACGGUAUGGUGACUUUUUAUGAUUUAAAGUCGUAAA--GUGCGA-AACGCCGGCAAGUAUUUACGACUCGGAAAUGGAAAUAUCAUGCCGUUGGGUAAGUAC--CCCAGCUGACUGGCU----UCGAGCUCUCC------ ((((((.((((((((((((((((..(((((((..(((..(((((((((--.(((..-.......)))....)))))))))..)))))))))))))))))))).)))))).)).)--)))((((((......----)).))))....------ ( -49.90, z-score = -3.11, R) >droPer1.super_9 1454699 137 + 3637205 GGGGGCAUGCCUACGGUAUGGUGACUUUUUAUGAUUUAAAGUCGUAAA--GUGCGA-AACGCCGGCAAGUAUUUACGACUCGGAAAUGGAAAUAUCAUGCCGUUGGGUAAGUAC--CCCAGCUGACUGACU----CCGAGCUCUCC------ ((((((.((((((((((((((((..(((((((..(((..(((((((((--.(((..-.......)))....)))))))))..)))))))))))))))))))).)))))).)).)--)))(((((.......----.).))))....------ ( -48.60, z-score = -2.93, R) >droWil1.scaffold_180949 2348895 127 - 6375548 --------CAGGACAAG-UGAUGGCUUUUUAUGAUUUAAAGUCAUAAAAUGUGCGACGACGCCAGCAAAUAUUUACGACUCGGAAAUGGAAAUGGCAUGCCAUUUGGUAAGUAC--CCAAACCGGCUGUGU----CUUAACC---------- --------.((((((.(-(..(.((.(((((((((.....)))))))))...)).)..))((((.....(((((.((...)).)))))....))))..(((.(((((.......--)))))..)))..)))----)))....---------- ( -31.90, z-score = -0.88, R) >droVir3.scaffold_13049 7737188 116 + 25233164 --------AUAAGCGAUGUA-UAUUUUUUUAUGAUUUAAAGUCGUAAA-UUUGUGG------CAGCAUAUAUUUACGACUC-------GAA------UGCCAUUUGGUAAGUACCCGGGAG----CCGGAUGUCCCCGGAUACGGCCUG--- --------....((......-..................(((((((((-(.((((.------...)))).)))))))))).-------...------((((....)))).(((.(((((..----(.....)..))))).))).))...--- ( -32.70, z-score = -1.55, R) >droMoj3.scaffold_6680 5984863 125 + 24764193 --------AUAAGCGAUGUA-U-UCCUUUUAUGAUUUAAAGUCGUAAA-AGUGCGG------CAGCAUAUAUUUACGACUCAGAAAUGGAA------UGCCAUUGAGUAAGUACCCAAGAG----CCGACUGUCCCCGGAUGCGGGCCAUUG --------.((..(((((((-(-(((.(((.(((......((((((((-.((((..------..))))...))))))))))).))).))))------)).)))))..))....(((.....----(((........)))....)))...... ( -33.30, z-score = -1.68, R) >droGri2.scaffold_15110 19419046 127 - 24565398 --------AUAAGCAAUGU----UUUUUUUAUGAUUUAAAGUCGUAAA-UUUGUGG------CAGCAUAUAUUUACGACUCAGAAAUGGAA------UGCCAUUCGAUAAGUACCCGGGAGGCGUCCACGGAUUCCCGAAUUCGGAAACACA --------........(((----((((............(((((((((-(.((((.------...)))).))))))))))..(((.((((.------((((.((((.........)))).))))))))(((....)))..)))))))))).. ( -34.30, z-score = -2.10, R) >consensus ________GGGGACAAC_CGAUGACUUUUUAUGAUUUAAAGUCGUAAA__GUGCGG_GGAGCCAGCAAGUAUUUACGACUCGGAAAUGGAAAUAUCAUGCCAUUUGGUAAGUAC__CCCAGCCAGCUGACU____UUGGAUUCGGC______ ............((............((((((..(((..(((((((((....((..........)).....)))))))))..)))))))))......((((....)))).))........................................ (-12.64 = -13.24 + 0.60)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:20:55 2011