
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 5,036,279 – 5,036,379 |
| Length | 100 |
| Max. P | 0.999771 |

| Location | 5,036,279 – 5,036,379 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 74.96 |
| Shannon entropy | 0.39292 |
| G+C content | 0.47756 |
| Mean single sequence MFE | -39.41 |
| Consensus MFE | -28.31 |
| Energy contribution | -27.93 |
| Covariance contribution | -0.37 |
| Combinations/Pair | 1.13 |
| Mean z-score | -4.61 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.35 |
| SVM RNA-class probability | 0.999771 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
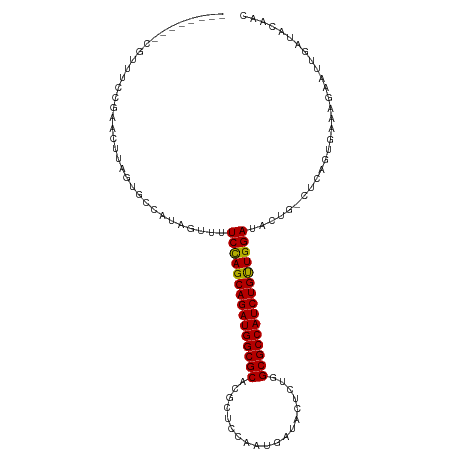
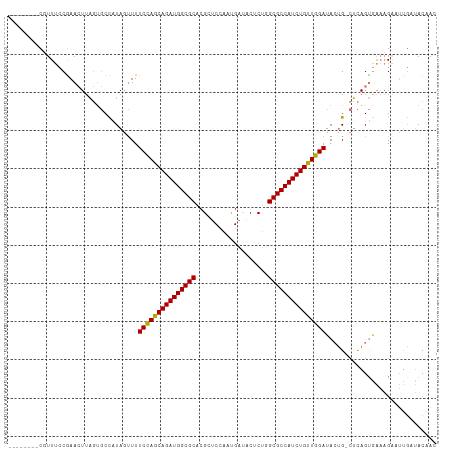
>dm3.chr2L 5036279 100 + 23011544 --------CGUUUCCGAAGUUU---CCCUAGCUUUCCAGCAGAUGGCGCACGUUCCAACGACACUCUUGCGCCAUCUGUUGGAUACUG-CGUAGCGAAAGAUUUGAUACAAU --------.((.((.((..(((---(.(((((..(((((((((((((((((((....))).......))))))))))))))))....)-).))).))))..)).)).))... ( -39.21, z-score = -5.38, R) >droEre2.scaffold_4929 5115271 109 + 26641161 CUUUUCUAAGUUGGCAAACUUAGUGCUAUAAUUUUCUAGCAGAUGGCGCACG--CAGAUGAUACACGUGCGCCAUCUGCUGGAUACAG-CUCAGUACUCGGAACUUGACAAC .........((((.(((.((.((((((.......((((((((((((((((((--...........)))))))))))))))))).....-...)))))).))...))).)))) ( -45.26, z-score = -6.12, R) >droSec1.super_5 3122795 104 + 5866729 --------CUUUUCCGAACUUAGUGCCAUAGUUUUCCAGCAGAUGGCGCACGCUCCAAUGAUACUCUGGCGCCAUCUGUUGGAUAGUGGCUCAGUGAAAGACUUGCUACAAC --------..............(.(((((.....(((((((((((((((.((..............)))))))))))))))))..))))).)((..(.....)..))..... ( -36.84, z-score = -3.20, R) >droSim1.chr2L_random 279557 104 + 909653 --------CUUUUCCGAACUUAGUGCCAUAGUUUUCCAGCAGAUGGCGCACGCUCCAAUGAUACUCUGGCGCCAUCUGUUGGAUACUGGCUCAGUGAAAGAAUUGAUACAAC --------................((((..((..(((((((((((((((.((..............))))))))))))))))).))))))(((((......)))))...... ( -36.34, z-score = -3.74, R) >consensus ________CGUUUCCGAACUUAGUGCCAUAGUUUUCCAGCAGAUGGCGCACGCUCCAAUGAUACUCUGGCGCCAUCUGUUGGAUACUG_CUCAGUGAAAGAAUUGAUACAAC ..................................(((((((((((((((...................)))))))))))))))............................. (-28.31 = -27.93 + -0.37)

| Location | 5,036,279 – 5,036,379 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 74.96 |
| Shannon entropy | 0.39292 |
| G+C content | 0.47756 |
| Mean single sequence MFE | -36.88 |
| Consensus MFE | -22.27 |
| Energy contribution | -23.28 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.30 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.25 |
| SVM RNA-class probability | 0.998064 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
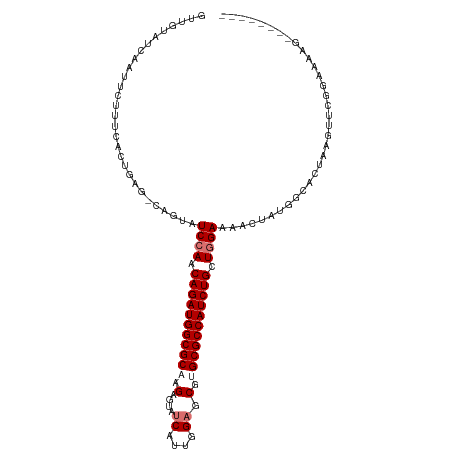
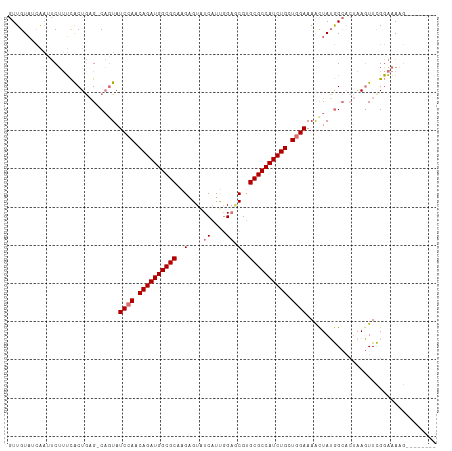
>dm3.chr2L 5036279 100 - 23011544 AUUGUAUCAAAUCUUUCGCUACG-CAGUAUCCAACAGAUGGCGCAAGAGUGUCGUUGGAACGUGCGCCAUCUGCUGGAAAGCUAGGG---AAACUUCGGAAACG-------- ...((.((..(..((((.(((.(-(....((((.(((((((((((...((.((....)))).))))))))))).))))..))))).)---)))..)..)).)).-------- ( -37.50, z-score = -3.61, R) >droEre2.scaffold_4929 5115271 109 - 26641161 GUUGUCAAGUUCCGAGUACUGAG-CUGUAUCCAGCAGAUGGCGCACGUGUAUCAUCUG--CGUGCGCCAUCUGCUAGAAAAUUAUAGCACUAAGUUUGCCAACUUAGAAAAG ....(((.((.......)))))(-(((((((.((((((((((((((((.........)--))))))))))))))).))....)))))).(((((((....)))))))..... ( -45.10, z-score = -5.97, R) >droSec1.super_5 3122795 104 - 5866729 GUUGUAGCAAGUCUUUCACUGAGCCACUAUCCAACAGAUGGCGCCAGAGUAUCAUUGGAGCGUGCGCCAUCUGCUGGAAAACUAUGGCACUAAGUUCGGAAAAG-------- ((..(((...(.(((.....))).)....((((.((((((((((....((.((....))))..)))))))))).))))...)))..))................-------- ( -31.70, z-score = -1.32, R) >droSim1.chr2L_random 279557 104 - 909653 GUUGUAUCAAUUCUUUCACUGAGCCAGUAUCCAACAGAUGGCGCCAGAGUAUCAUUGGAGCGUGCGCCAUCUGCUGGAAAACUAUGGCACUAAGUUCGGAAAAG-------- ..........((((...(((..((((.(.((((.((((((((((....((.((....))))..)))))))))).)))).)....))))....)))..))))...-------- ( -33.20, z-score = -2.30, R) >consensus GUUGUAUCAAUUCUUUCACUGAG_CAGUAUCCAACAGAUGGCGCAAGAGUAUCAUUGGAGCGUGCGCCAUCUGCUGGAAAACUAUGGCACUAAGUUCGGAAAAG________ .............................((((.(((((((((((.(....((....)).).))))))))))).)))).................................. (-22.27 = -23.28 + 1.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:17:50 2011