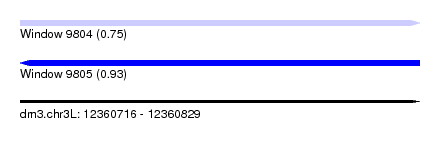
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 12,360,716 – 12,360,829 |
| Length | 113 |
| Max. P | 0.930967 |

| Location | 12,360,716 – 12,360,829 |
|---|---|
| Length | 113 |
| Sequences | 7 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 69.16 |
| Shannon entropy | 0.59546 |
| G+C content | 0.35153 |
| Mean single sequence MFE | -20.79 |
| Consensus MFE | -6.67 |
| Energy contribution | -7.57 |
| Covariance contribution | 0.90 |
| Combinations/Pair | 1.50 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.32 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.754229 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
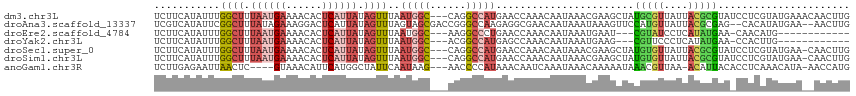
>dm3.chr3L 12360716 113 + 24543557 UCUUCAUAUUUGGCUUUAAUGAAAACACUCAUUAUAGUUUAAUGGC---CAGGCCAUGAACCAAACAAUAAACGAAGCUAUGCGUUAUUACGCGUAUCCUCGUAUGAAACAACUUG ..(((((.(((((((.((((((......)))))).))....(((((---...)))))...)))))......((((.(.(((((((....))))))).).)))))))))........ ( -28.10, z-score = -3.63, R) >droAna3.scaffold_13337 22002733 112 - 23293914 UCGUCAUAUUCGGCUUUAUAGAAAGGACUCAUUAUAGUUUAGUAGCGACCGGGCCAAGAGGCGAACAAUAAAUAAAGUUCCAUGUUAUUACGCGAG--CACAUAUGAA--AACUUG ...((((((...((((....((......))......((.((((((((.....(((....)))((((..........))))..)))))))).)))))--)..)))))).--...... ( -21.80, z-score = -0.39, R) >droEre2.scaffold_4784 12355514 97 + 25762168 UCUUCAUAUUUGGCUUUAAUGAAAACACUCAUUAUAGUUUAAUGGC---AAGGCCCUGAACCAAACAAUAAAUGAAU---CGUAUCCUCAUAUGAA-CAACAUG------------ ..((((((((..(((.((((((......)))))).)))..)))(((---...)))................)))))(---(((((....)))))).-.......------------ ( -14.60, z-score = -0.83, R) >droYak2.chr3L 12415627 97 + 24197627 UCUUCAUAUUUGGCUUUAAUGAAAACACUCAUUAUAGUUUAAUGGC---ACGGCCAUGAGCCAAACAAUAAAUGAAG---CGUUCCCUCAUAUGAA-CCACUUG------------ .((((((.((((((((((((((......)))))).......(((((---...)))))))))))))......))))))---.((((........)))-)......------------ ( -21.90, z-score = -2.74, R) >droSec1.super_0 4544767 112 + 21120651 UCUUCAUAUUUGGCUUUAAUGAAAACACUCAUUAUAGUUUAAUGGC---CAGGCCAUGAACCAAACAAUAAACGAAGCUAUGUGUUAUUACGCGUAUCCUCGUAUGAA-CAACUUG ..(((((.(((((((.((((((......)))))).))....(((((---...)))))...)))))......((((.(.(((((((....))))))).).)))))))))-....... ( -25.90, z-score = -2.83, R) >droSim1.chr3L 11738925 112 + 22553184 UCUUCAUAUUUGGCUUUAAUGAAAACACUCAUUAUAGUUUAAUGGC---CAGGCCAUGAACCAAACAAUAAACGAAGCUAUGUGUUAUUACGCGUAUCCUCGUAUGAA-CAACUUG ..(((((.(((((((.((((((......)))))).))....(((((---...)))))...)))))......((((.(.(((((((....))))))).).)))))))))-....... ( -25.90, z-score = -2.83, R) >anoGam1.chr3R 52337259 107 + 53272125 UCUUGAGAAUUAACUC----GUAAACAUUCAUGGCUAUUCAAUAAG---AACCCCAUAAACAAUCAAAUAAACAAAAAUAAACGUUAA-ACAUUACACCUCAAACAUA-AACCAUG ..(((((.........----..........((((...(((.....)---))..))))..........................((...-.....))..))))).....-....... ( -7.30, z-score = -1.05, R) >consensus UCUUCAUAUUUGGCUUUAAUGAAAACACUCAUUAUAGUUUAAUGGC___CAGGCCAUGAACCAAACAAUAAACGAAGCUACGUGUUAUUACGCGAA_CCUCAUAUGAA__AACUUG ...........((((.((((((......)))))).))))..(((((......))))).......................(((((....)))))...................... ( -6.67 = -7.57 + 0.90)


| Location | 12,360,716 – 12,360,829 |
|---|---|
| Length | 113 |
| Sequences | 7 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 69.16 |
| Shannon entropy | 0.59546 |
| G+C content | 0.35153 |
| Mean single sequence MFE | -23.81 |
| Consensus MFE | -13.43 |
| Energy contribution | -13.50 |
| Covariance contribution | 0.07 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.930967 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
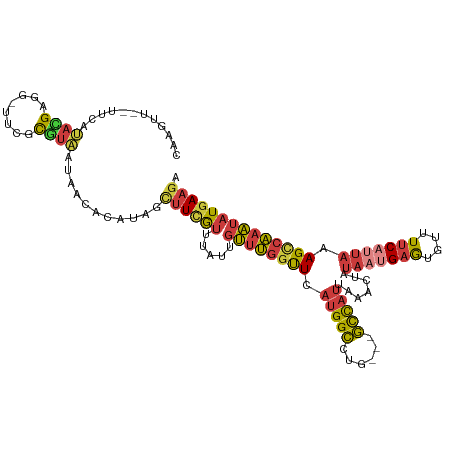
>dm3.chr3L 12360716 113 - 24543557 CAAGUUGUUUCAUACGAGGAUACGCGUAAUAACGCAUAGCUUCGUUUAUUGUUUGGUUCAUGGCCUG---GCCAUUAAACUAUAAUGAGUGUUUUCAUUAAAGCCAAAUAUGAAGA .......((((((((((((.((.((((....)))).)).)))))).....((((((((.(((((...---))))).......(((((((....))))))).)))))))))))))). ( -30.90, z-score = -2.68, R) >droAna3.scaffold_13337 22002733 112 + 23293914 CAAGUU--UUCAUAUGUG--CUCGCGUAAUAACAUGGAACUUUAUUUAUUGUUCGCCUCUUGGCCCGGUCGCUACUAAACUAUAAUGAGUCCUUUCUAUAAAGCCGAAUAUGACGA ......--.((((((..(--((...((....))((((((....(((((((((..(((....)))..(((....))).....)))))))))...))))))..)))...))))))... ( -17.40, z-score = 0.94, R) >droEre2.scaffold_4784 12355514 97 - 25762168 ------------CAUGUUG-UUCAUAUGAGGAUACG---AUUCAUUUAUUGUUUGGUUCAGGGCCUU---GCCAUUAAACUAUAAUGAGUGUUUUCAUUAAAGCCAAAUAUGAAGA ------------(((((((-((...((((((((((.---..(((((....(((((((....(((...---))))))))))...)))))))))))))))...)))..)))))).... ( -21.90, z-score = -1.30, R) >droYak2.chr3L 12415627 97 - 24197627 ------------CAAGUGG-UUCAUAUGAGGGAACG---CUUCAUUUAUUGUUUGGCUCAUGGCCGU---GCCAUUAAACUAUAAUGAGUGUUUUCAUUAAAGCCAAAUAUGAAGA ------------......(-(((........)))).---((((((.....((((((((.(((((...---))))).......(((((((....))))))).)))))))))))))). ( -25.90, z-score = -2.00, R) >droSec1.super_0 4544767 112 - 21120651 CAAGUUG-UUCAUACGAGGAUACGCGUAAUAACACAUAGCUUCGUUUAUUGUUUGGUUCAUGGCCUG---GCCAUUAAACUAUAAUGAGUGUUUUCAUUAAAGCCAAAUAUGAAGA ...((((-(...((((.(....).)))))))))......((((((.....((((((((.(((((...---))))).......(((((((....))))))).)))))))))))))). ( -26.10, z-score = -1.43, R) >droSim1.chr3L 11738925 112 - 22553184 CAAGUUG-UUCAUACGAGGAUACGCGUAAUAACACAUAGCUUCGUUUAUUGUUUGGUUCAUGGCCUG---GCCAUUAAACUAUAAUGAGUGUUUUCAUUAAAGCCAAAUAUGAAGA ...((((-(...((((.(....).)))))))))......((((((.....((((((((.(((((...---))))).......(((((((....))))))).)))))))))))))). ( -26.10, z-score = -1.43, R) >anoGam1.chr3R 52337259 107 - 53272125 CAUGGUU-UAUGUUUGAGGUGUAAUGU-UUAACGUUUAUUUUUGUUUAUUUGAUUGUUUAUGGGGUU---CUUAUUGAAUAGCCAUGAAUGUUUAC----GAGUUAAUUCUCAAGA .......-....(((((((..((((..-..((((........))))....(((..((((((((.(((---(.....))))..))))))))..))).----..))))..))))))). ( -18.40, z-score = -0.61, R) >consensus CAAGUU__UUCAUACGAGG_UUCGCGUAAUAACACAUAGCUUCGUUUAUUGUUUGGUUCAUGGCCUG___GCCAUUAAACUAUAAUGAGUGUUUUCAUUAAAGCCAAAUAUGAAGA ............((((........))))...........((((((.....((((((((.(((((......))))).......(((((((....))))))).)))))))))))))). (-13.43 = -13.50 + 0.07)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:20:52 2011